0305
Accelerated respiratory-resolved 4D-MRI with separable spatio-temporal neural networks1Deparment of Radiotherapy, University Medical Center Utrecht, Utrecht, Netherlands, 2Computational Imaging Group for MR Diagnostics & Therapy, University Medical Center Utrecht, Utrecht, Netherlands
Synopsis
Four-dimensional (4D) respiratory-resolved imaging is crucial for managing respiratory motion in radiotherapy, enabling irradiation of highly mobile tumours. However, acquiring high-quality 4D-MRI requires long acquisition (typically ≥5 minutes) and long iterative reconstructions, limiting treatment efficiency. Recently, deep learning has been proposed to accelerate undersampled MRI reconstruction. However, it has not been established whether deep learning may reconstruct high-quality 4D-MRI from accelerated acquisitions. This work proposes a small deep learning model that exploits the spatio-temporal information present in 4D-MRI, allowing to split the reconstruction into two separated branches, enabling high-quality retrospectively-accelerated 4D-MRI acquired in ~60 seconds and reconstructed in 16 seconds.
Introduction
Four-dimensional (4D) respiratory-resolved imaging is crucial for managing respiratory motion in radiotherapy, enabling accurate irradiation of highly mobile tumours1, and facilitating treatment adaptation based on daily motion2.However, obtaining high-quality 4D-MRI requires long acquisition (typically ≥5 minutes) and long iterative reconstructions limiting treatment efficiency.
Recently, deep learning (DL) has been proposed to accelerate undersampled MRI reconstruction3, demonstrating its potential also for 4D-MRI4. However, it has not been established whether DL may reconstruct high-quality 4D-MRI from accelerated acquisitions.
Reconstructing complete 4D-MRI volumes with a single model requires performing 4D convolutions, which is an expensive operation that may result in an overly large model, e.g., >100M trainable parameters. Training such large models requires specialized hardware and large training datasets while being prone to overfitting.
Previous research5,6,7 showed that separating spatial and temporal information is a powerful technique, leading to efficient DL architectures. We propose using a model that exploits the spatio-temporal information present in 4D-MRI, allowing to split the reconstruction into two separated branches. Also, we perform 2D convolutions rather than training a single model that performs 3D convolutions.
Such models have few parameters, leading to low memory usage, fast inference, and low risk of overfitting. We investigate the performance of 2D+t models, studying whether they may enable high-quality accelerated 4D-MRI acquisition and reconstruction.
Materials & Methods
Data preparationTwenty-seven patients with lung cancer included in an IRB-approved study were imaged in free-breathing during Gadolinium injection on a 1.5T MRI (MR-RT, Philips Healthcare) using a fat-suppressed golden-angle radial stack-of-stars T1-w GRE thorax MRI for approximately five minutes (resolution=2.2x2.2x3.5mm3, FOV=350x350x270mm3, slice direction=feet-head). K-space was retrospectively sorted in 10 respiratory bins based on the relative amplitude of the self-navigation signal present in radial k-space8. Complex XD-GRASP9 reconstructions with total variation over the respiratory phases ($$$\lambda=0.03$$$) served as the target image. Accelerated complex NUFFT-adjoint images were reconstructed10 by undersampling the sorted bins with acceleration factors R4D=1, 2, 4, 8, and 10, simulating an acquisition of approximately Tacq=300, 150, 60, 30, and 25 seconds long, respectively. This corresponded to an undersampling factor per bin of RNyquist = 4, 8, 16, 32, and 40, respectively.
For evaluation purposes, we computed deformation vectors fields (DVFs) between every respiratory phase and the end-exhale state using a free-form deformation algorithm11.
Model architecture
The proposed model will reconstruct a 2D+t respiratory-resolved volume $$$y$$$ with $$$n_p$$$ phases using a spatial and a temporal branch. The spatial branch learns a volume $$$\Psi$$$ by performing $$$1\times c_x \times c_y$$$ complex convolutions, while the temporal branch learns a volume $$$\Xi$$$ by performing $$$n_p\times1\times1$$$ complex convolutions. The 4D reconstruction is then achieved by learning $$$y_{t+1}=y_t+\Psi\Xi$$$ for every slice (Figure 1).
We implemented an iterative model12 with residual connections between $$$y_{t+1}$$$ and $$$y_{t}$$$, using four iterations to learn complex XD-GRASP reconstructed slices from complex NUFFT-reconstructed slices of 4D-MRI. Each iteration used a spatial and a temporal branch with five complex convolution layers, employing a cardioid non-linear activation function13 ($$$c_x=c_y=3$$$). In total, the model has approximately 238,000 trainable parameters.
Training and experiments
Ten models with identical architectures were trained on axial and sagittal slices, one for every undersampling factor R4D. Every model was trained on 17 patients, evaluated on five, and tested on five other patients. Models were optimized using the AdamW optimizer (lr=10-3, weight-decay=10-4) with a batch size of 3, minimizing the hybrid magnitude SSIM/$$$\perp$$$-Loss14 for 20 epochs.
Evaluation
Models were evaluated on all patients in the test set using the magnitude SSIM of the full 4D reconstruction compared to the XD-GRASP reconstruction and the end-point error (EPE) of DVFs obtained with the free-form deformation against the XD-GRASP DVFs. Statistical significance (p<0.05) was established using a paired t-test.
Results and discussion
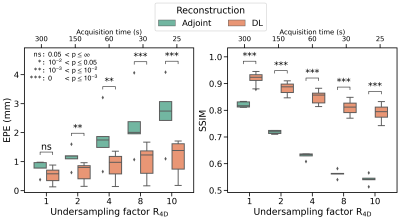
The models were trained in ~6 hours using ~18GB memory, and inference took ~16 seconds for full 4D-MRI.Using the proposed deep learning model significantly improved image quality (Figure 2), demonstrating that spatio-temporally decomposed models can effectively reconstruct high-quality 4D-MRI. Compared to XD-GRASP, the average SSIM of non-accelerated 4D-MRI (R4D=1) increased from $$$0.82\pm0.01$$$ ($$$\mu\pm\sigma$$$) using NUFFT reconstructions to $$$0.92\pm0.02$$$ with our model. Image quality is significantly increased for accelerated reconstructions (Figure 3). For R4D=4, the acquisition time decreased to ~60s, while the mean EPE decreased from $$$1.80\pm0.92$$$ mm for NUFFT reconstructions to $$$0.85\pm0.43$$$ mm (Figure 4), although blurring can be observed in the DL reconstructions.
Using R4D>4 reduces reconstructed image quality and shows decreased motion magnitude. However, considering an EPE<1mm as clinically acceptable, reconstructions up to R4D=4 may be justified, allowing for accurate motion estimation with a retrospective acquisition time Tacq$$$\approx$$$60s.
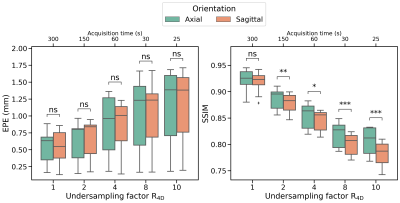
No significant differences were found between axial and sagittal reconstructions for motion estimation. However, models evaluated on axial images show higher image reconstruction quality than models evaluated on sagittal images (Figure 5).
Future work will investigate model performance on prospectively undersampled MRI and comparison to not spatio-temporally decomposed models.
Using high-quality accelerated 4D-MRI could shorten the acquisition time, increasing patient comfort and improving treatment throughput. Moreover, it could enable accelerated high-quality time-resolved imaging15.
Conclusion
We have presented a small deep learning model for 4D-MRI reconstruction that splits the spatio-temporal information into two separated branches. Retrospectively-accelerated 4D-MRI (Tacq$$$\approx$$$60s) was reconstructed in 16s with high quality, enabling accurate motion estimation compared to XD-GRASP (EPE<1mm).Acknowledgements
This work is part of the research program HTSM with project number 15354, which is (partly) financed by the Netherlands Organisation for Scientific Research (NWO) and Philips Healthcare. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Quadro RTX 5000 GPU used for prototyping this research.References
[1] Korreman et al. "Image-guided radiotherapy and motion management in lung cancer" Br J Radiol. 2015
[2] Stemkens et al. "Nuts and bolts of 4D-MRI for radiotherapy." PMB 63.21 (2018)
[3] Hammernik et al. "Learning a variational network for reconstruction of accelerated MRI data" MRM 79.6 (2017)
[4] Freedman et al. "Rapid 4D-MRI reconstruction using a deep radial convolutional neural network: Dracula." Radiotherapy and Oncology 159 (2021)
[5] Huttinga et al. "Prospective 3D+t non-rigid motion estimation at high frame-rate from highly undersampled k-space data: validation and preliminary in-vivo results" Proc. Intl. Soc. Mag. Reson. Med. 27 (2019)
[6] Kofler et al. "Spatio-temporal deep learning-based undersampling artefact reduction for 2D radial cine MRI with limited training data" IEEE TMI 39.3 (2019)
[7] Küstner, Thomas, et al. "CINENet: deep learning-based 3D cardiac CINE MRI reconstruction with multi-coil complex-valued 4D spatio-temporal convolutions." Scientific reports 10.1 (2020)
[8] Stemkens et al. "Optimizing 4-dimensional magnetic resonance imaging data sampling for respiratory motion analysis of pancreatic tumors" IJROBP 91.3 (2015)
[9] Feng et al. "XD‐GRASP: golden‐angle radial MRI with reconstruction of extra motion‐state dimensions using compressed sensing." MRM 75.2 (2016)
[10] Fessler and Sutton. "Nonuniform fast Fourier transforms using min-max interpolation." IEEE transactions on signal processing 51.2 (2003)
[11] Beare et al. “Image Segmentation, Registration and Characterization in R with SimpleITK”, J Stat Softw 86.8 (2018)
[12] Adler et al. "Solving ill-posed inverse problems using iterative deep neural networks" IP 33.12 (2017)
[13] Virtue et al. "Better than real: Complex-valued neural nets for MRI fingerprinting." IEEE ICIP (2017)
[14] Terpstra et al. "Rethinking complex image reconstruction: ⟂-loss for improved complex image reconstruction with deep learning" Proc. Intl. Soc. Mag. Reson. Med. 29 (2021)
[15] Feng et al. "MRSIGMA: Magnetic Resonance SIGnature MAtching for real-time volumetric imaging" MRM 84.3 (2020)
Figures
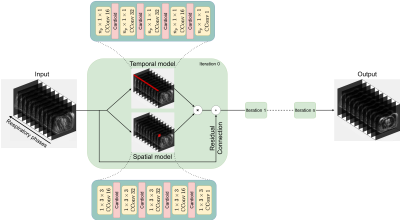
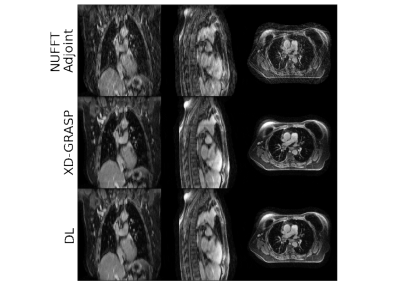
Figure 2: Example reconstruction with R4D=1. Here, a patient (female, 71 years old, T3N0M0 squamous cell-carcinoma) from the test set is shown. Top row shows the NUFFT-adjoint reconstruction, middle row the XD-GRASP reconstruction, and the bottom row shows the deep learning reconstruction. The acquisition time was approximately 5 minutes. Average SSIM increased from 0.81 using NUFFT adjoint reconstructions to 0.94 with DL reconstructions compared to XD-GRASP, while the EPE decreased from 0.98 mm to 0.38 mm.
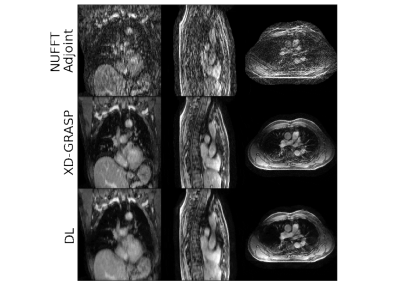
Figure 3: Example reconstruction with R4D=4. Here, a patient (male, 72 years old, T4N1M0 small-cell carcinoma) from the test set is shown. Top row shows the NUFFT-adjoint reconstruction, middle row the XD-GRASP reconstruction, and the bottom row shows the deep learning reconstruction. The acquisition time was approximately 1 minutes. Average SSIM increased from 0.63 using a NUFFT-adjoint reconstruction to 0.87 with DL reconstructions compared to XD-GRASP, while the EPE decreased from 1.87 mm to 1.10 mm.