0299
The deep SECRET to accelerated first-pass perfusion cardiac MRI
Elena Martín-González1, Ebraham Alskaf2, Amedeo Chiribiri 2, Pablo Casaseca-de-la-Higuera1, Carlos Alberola-López1, Rita G. Nunes3,4, and Teresa Correia2,5
1Laboratorio de Procesado de Imagen, Universidad de Valladolid, Valladolid, Spain, 2School of Biomedical Engineering and Imaging Sciences, King’s College London, London, United Kingdom, 3Institute for Systems and Robotics, Lisbon, Portugal, 4Department of Bioengineering, Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal, 5Centre for Marine Sciences - CCMAR, Faro, Portugal
1Laboratorio de Procesado de Imagen, Universidad de Valladolid, Valladolid, Spain, 2School of Biomedical Engineering and Imaging Sciences, King’s College London, London, United Kingdom, 3Institute for Systems and Robotics, Lisbon, Portugal, 4Department of Bioengineering, Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal, 5Centre for Marine Sciences - CCMAR, Faro, Portugal
Synopsis
First-pass perfusion cardiac magnetic resonance (FPP-CMR) is becoming essential to detect blow flow anomalies. However, the need for real-time acquisitions limits the achievable spatial resolution and coverage of the heart. To keep both within a reasonable range, FPP-CMR needs to be accelerated. A SElf-Supervised aCcelerated REconsTruction (SECRET) DL framework is presented to speed-up reconstruction of FPP-CMR images from undersampled (k,t)-space data. The physical reconstruction models are used to train deep neural networks without requiring fully sampled images. SECRET achieves good quality reconstructions at a variety of acceleration rates, with significant speed-ups compared to the state-of-the-art.
Introduction
Coronary artery disease (CAD) is the occlusion of coronary arteries that causes abnormalities in blood flow to the heart. First-pass perfusion cardiac magnetic resonance (FPP-CMR) is rapidly evolving into an essential tool to detect blood flow anomalies1-4. However, FPP-CMR time frames must be acquired in real-time to capture the passage of a contrast agent bolus through the heart, and hence, spatial resolution and coverage of the heart are compromised. Undersampled reconstruction methods have been proposed to accelerate FPP-CMR acquisitions in order to improve both resolution and coverage5-7. However, these methods can lead to long reconstruction times. This work aims to speed up reconstructions by obtaining the contrast-enhanced dynamic image series from undersampled FPP-CMR using a deep learning (DL) based reconstruction method. These images are then used to generate quantitative perfusion maps using a tracer kinetic model8-10. DL has already been applied to MR image reconstruction, specifically on knee11-13, brain11-14 and cardiac15-16 MR imaging, using both supervised11-12,14 and self-supervised learning13,17. Occasionally, the network is unrolled to emulate compressed sensing (CS) iterative reconstruction, giving rise to a cascade of convolutional neural networks (CNNs)11-12,16. Supervised learning techniques need fully sampled reference images to train the network, which are not available in FPP-CMR, particularly at high spatial resolutions. To overcome this limitation, a SElf-Supervised aCcelerated REconsTruction (SECRET) DL framework is proposed to reconstruct FPP-CMR images from undersampled (k,t)-space data.Methods
Acquisition: Rest and stress FPP-CMR acquisitions were performed in 21 patients using a single-bolus injection of 0.05 mmol/kg Gadobutrol and a 1.5T scanner (Siemens MAGNETOM Aera). A free-breathing FLASH perfusion dual-sequence10 was used to acquire a low-resolution arterial input function image and three short-axis slices (basal, mid and apical) high-resolution myocardial images using the following parameters: FOV=340x308mm2, in-plane resolution=2.2x2.2mm2, slice thickness=10mm, TR/TE=2.1/1ms, flip angle=8º, parallel imaging acceleration 3, saturation recovery time=100ms, total scan duration=60s. Undersampled datasets were generated for 3x, 6x and 10x acceleration factors using radial (k,t)-sampling.Preprocessing: Images were resized to a spatial resolution of 2x2mm2, the k-space padded to obtain an image of 256x256 pixels, each slice was interpolated to a fixed number of (60) frames, and image intensity normalisation was applied. Image pre-registration was also carried out to correct for respiratory motion.
Reconstruction and Analysis: The proposed SECRET method combines physics, U-NET18 and residual learning17 to reconstruct contrast-enhanced dynamic images from the undersampled (k,t)-space data (Fig.1). Considering only the undersampled data when enforcing data consistency, the network can be trained using the physical models in the reconstruction17, without requiring fully sampled images. The loss is computed with the estimated undersampled (k,t)-space and the input, to guide the training phase.
Images were reconstructed for 5 patients using SECRET, CS and MoDL11. MoDL uses supervised learning and an unrolled architecture for a number of iterations, K. Reconstructions were quantitatively evaluated against the fully-sampled images using normalized mean square error (NMSE), structural similarity (SSIM) index and peak signal-to-noise ratio (PSNR). Quantitative maps were generated using the Patlak model19.
Implementation: A 60-16-24 training-validation-test split was used. SECRET took ~30 min to train using the Adam optimizer20 and learning rate of 10-4. Training MoDL for K=1 took ~1h30min and for K=10 ~45h.
Results and Discussion
Figure 2 shows the SECRET reconstructions from one representative patient together with the reference, CS and MoDL (K=1) reconstructions. Although SECRET reconstructions are slightly blurred, due to residual learning from the average image (also blurred due to residual motion), there is a quality improvement compared to MoDL (trained for a similar amount of time).Quality metrics indicate that SECRET achieves a more consistent agreement with the reference as the acceleration factor increases than MoDL (Fig. 3). CS and MoDL (K=10) show the best agreement with the reference, but reconstructions take ~87.08s and ~1.99s, respectively, whereas MoDL (K=1) takes ~0.21s and SECRET only 0.15s.
SECRET does not include any explicit regularisation term. However, due to residual learning, all reconstructions provided by the framework are inherently corrected. Such good PSNR, SSIM and NRMSE values obtained when the reference images are affected by residual respiratory motion, would certainly improve if regularisation were added.
The SECRET quantitative maps are comparable to the reference maps, showing less blurring than MoDL maps (Fig. 5).
Conclusion
A physics-informed self-supervised deep learning reconstruction framework for accelerating FPP-CMR scans has been described. SECRET provides good quality reconstructions at all acceleration rates, without using fully sampled data for training. Our method shows promising results, with the potential for improvement coupled with explicit regularization and parameter optimisation, which will be explored in future work.Acknowledgements
This work is part of a project that has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 867450. Authors also thank European Social Fund, Operational Programme of Castilla y León, and the Junta de Castilla y León. This work has also been supported by Agencia Estatal de Investigación through grants TEC2017-82408-R and PID2020-115339RB-I00 and by Fundação para a Ciência e Tecnologia (FCT) through grant UIDP/50009/2020.References
- Heo, R., Nakazato, R., Kalra, D., Min, J.K.: Noninvasive imaging in coronary artery disease. In: Seminars in nuclear medicine. vol. 44, pp. 398–409. Elsevier (2014)
- Foley, J.R.J.: Cardiovascular Magnetic Resonance Imaging for the Investigation of Ischaemic Heart Disease. Ph.D. thesis, University of Leeds (2018)
- Hendel, R.C., Friedrich, M.G., Schulz-Menger, J., Zemmrich, C., Bengel, F., Berman, D.S., Camici, P.G., Flamm, S.D., Le Guludec, D., Kim, R., et al.: Cmr first-pass perfusion for suspected inducible myocardial ischemia. JACC: Cardiovascular imaging 9(11), 1338–1348 (2016)
- Schwitter, J., Wacker, C.M., Wilke, N., Al-Saadi, N., Sauer, E., Huettle, K., Sch¨onberg, S.O., Luchner, A., Strohm, O., Ahlstrom, H., et al.: Mr-impact ii: Magnetic resonance imaging for myocardial perfusion assessment in coronary artery disease trial: perfusion-cardiac magnetic resonance vs. single-photon emission computed tomography for the detection of coronary artery disease: a comparative multicentre, multivendor trial. European heart journal 34(10), 775–781 (2013)
- Lingala, S.G., Hu, Y., DiBella, E., Jacob, M.: Accelerated dynamic mri exploiting sparsity and low-rank structure: kt slr. IEEE transactions on medical imaging 30(5), 1042–1054 (2011)
- Otazo, R., Kim, D., Axel, L., Sodickson, D.K.: Combination of compressed sensing and parallel imaging for highly accelerated first-pass cardiac perfusion mri. Magnetic resonance in medicine 64(3), 767–776 (2010)
- Vitanis, V., Manka, R., Giese, D., Pedersen, H., Plein, S., Boesiger, P., Kozerke, S.: High resolution three-dimensional cardiac perfusion imaging using compartment-based k-t principal component analysis. Magnetic resonance in medicine 65(2), 575–587 (2011)
- Correia, T., Schneider, T., Chiribiri, A.: Model-based reconstruction for highly accelerated first-pass perfusion cardiac mri. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 514–522. Springer (2019)
- Hsu, L.Y., Jacobs, M., Benovoy, M., Ta, A.D., Conn, H.M., Winkler, S., Greve, A.M., Chen, M.Y., Shanbhag, S.M., Bandettini, W.P., et al.: Diagnostic performance of fully automated pixel-wise quantitative myocardial perfusion imaging by cardiovascular magnetic resonance. JACC: Cardiovascular Imaging 11(5), 697–707 (2018)
- Kellman, P., Hansen, M.S., Nielles-Vallespin, S., Nickander, J., Themudo, R., Ugander, M., Xue, H.: Myocardial perfusion cardiovascular magnetic resonance: optimized dual sequence and reconstruction for quantification. Journal of Cardiovascular Magnetic Resonance 19(1), 1–14 (2017)
- Aggarwal, H.K., Mani, M.P., Jacob, M.: Modl: Model-based deep learning architecture for inverse problems. IEEE transactions on medical imaging 38(2), 394–405 (2018)
- Kocanaogullari, D., Eksioglu, E.M.: Deep learning for mri reconstruction using a novel projection based cascaded network. In: 2019 IEEE 29th International Workshop on Machine Learning for Signal Processing (MLSP). pp. 1–6. IEEE (2019)
- Yaman, B., Hosseini, S.A.H., Moeller, S., Ellermann, J., Ugurbil, K., Akcakaya, M.: Self-supervised learning of physics-guided reconstruction neural networks without fully sampled reference data. Magnetic resonance in medicine 84(6), 3172–3191 (2020)
- Do, W.J., Seo, S., Han, Y., Ye, J.C., Choi, S.H., Park, S.H.: Reconstruction of multicontrast mr images through deep learning. Medical physics 47(3), 983–997 (2020)
- Huang, Q., Yang, D., Wu, P., Qu, H., Yi, J., Metaxas, D.: Mri reconstruction via cascaded channel-wise attention network. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). pp. 1622–1626. IEEE (2019)
- Schlemper, J., Caballero, J., Hajnal, J.V., Price, A.N., Rueckert, D.: A deep cascade of convolutional neural networks for dynamic mr image reconstruction. IEEE transactions on Medical Imaging 37(2), 491–503 (2017)
- Liu, F., Kijowski, R., El Fakhri, G., Feng, L.: Magnetic resonance parameter mapping using model-guided self-supervised deep learning. Magnetic Resonance in Medicine (2021)
- Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. pp. 234–241. Springer (2015)
- Patlak, C.S., Blasberg, R.G., Fenstermacher, J.D.: Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. Journal of CerebralBlood Flow & Metabolism 3(1), 1–7 (1983)
- Kingma, D.P., Ba, J.: Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Figures
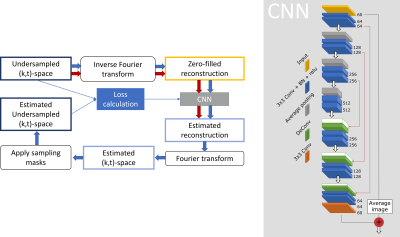
Figure 1. Flow chart of the proposed SECRET method for FPP-CMR. Blue arrows represent steps that take place during training, and red arrows represent the steps followed when making an estimation. The CNN is based on the U-Net18. Skip connections are included to maintain information from previous layers, and to avoid the problem of vanishing gradients during backpropagation. At the end of the CNN, residual learning has been appended17, adding the average image of the zero-filled reconstructions.
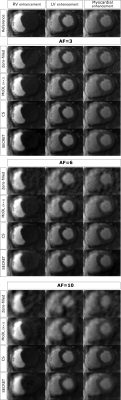
Figure 2. SECRET and MoDL (K=1) reconstructions obtained from 3x, 6x and 10x undersampled FPP-CMR data from one representative subject. Reference images are displayed for comparison. The right ventricle (RV), left ventricle (LV) and myocardial enhancement time frames are shown for one short axis slice. SECRET provides good quality images in a few seconds whereas CS takes about 1.5 min. SECRET requires shorter training times than MoDL and provides images that are visually better than those generated by MoDL.
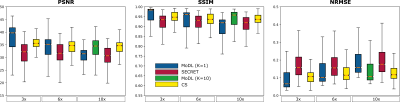
Figure 3. PSNR, SSIM and NRMSE between the reference and reconstructions obtained with SECRET, MoDL and CS methods, for 3x, 6x and 10x acceleration factors, for all patients. While the performance of MoDL becomes noticeably worse as the acceleration rate increases, SECRET maintains good image quality even at high acceleration rates. CS and MoDL (K=10) show the best agreement with the reference, but reconstructions (and MoDL training) take longer than MoDL (K=1) and SECRET.
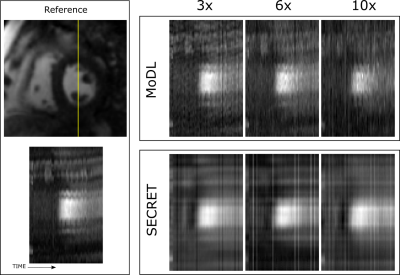
Figure 4. Representative 1D profiles obtained from MoDL and SECRET reconstructions. The profiles correspond to the location of the line in the reference image. Note that although the images have been pre-registered, there is still some residual motion. However, SECRET reconstructions are inherently motion corrected due to the residual learning.
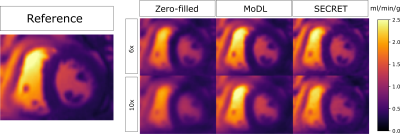
Figure 5. Quantitative maps (KTrans) obtained from 6x and 10x undersampled data using MoDL and SECRET. The reference image is displayed for comparison.
DOI: https://doi.org/10.58530/2022/0299