0261
Towards whole brain layer-fMRI connectivity: methodological advancements for functional layer connectomics1The Faculty of Psychology and Neuroscience, Maastricht University, Maastricht, Netherlands, 2Graduate School of Informatics and Engineering, The University of Electro-Communications, Tokyo, Japan, 3Scannexus, Maastricht, Netherlands, 4Brain Innovation, Maastricht, Netherlands, 5Center for Neuroscience and Biomedical Engineering, The University of Electro-Communications, Tokyo, Japan
Synopsis
Laminar-specific fMRI allows neuroscientists to address research questions of directional functional connectivity within and across brain areas. While recent sequence developments allow improvements in coverage and mitigations of venous biases, previous attempts of whole-brain connectome datasets turned out to be too artifact-dominated (Mueller 2021) to be neuroscientifically applicable. Here we present a new and improved sequence used for acquiring a relatively large open dataset of whole-brain laminar connectivity. Its purpose is to:
- investigate the reproducibility of laminar connectivity results,
- to benchmark and develop processing pipelines,
- and to explore which type of new neuroscientific research questions become addressable with laminar fMRI.
Purpose
Laminar-specific fMRI allows neuroscientists to address questions of directional functional connectivity within and across brain areas. While recent sequence developments allow improvements in coverage and mitigations of venous biases, previous attempts of whole-brain connectome datasets turned out to be too artifact-dominated (Mueller 2021) to be neuroscientifically applicable.Here, we describe our efforts to achieve a new and improved sequence for whole-brain layer-dependent functional connectome mapping. The developed methodology is used to acquire a large openly available dataset of cerebral blood volume and BOLD contrast. The purpose of this public dataset is multifold:
- At last year's ISMRM, we proposed a whole-brain protocol (https://cds.ismrm.org/protected/21MPresentations/abstracts/0630.html), which turned out to be unusable due to physiological and scanner artifacts (inflow and traveling wave artifact with long TRs Fig. 1). In this year's work, we aim to present solutions to those artifacts.
- We seek to provide a test bed for developing and benchmarking new layer-dependent preprocessing and analysis tools.
- We aim to characterise the layer-dependent signatures of common connectivity networks (Pais-Roldán, 2020) and their replicability.
- We aim to provide at least 50 runs of movie watching clips (14min each) to quantify reliability of laminar connectivity results.
- We want to explore which type of new neuroscientific research questions will become addressable with laminar fMRI.
Method
Scanning was performed on a SIEMENS MAGNETOM “Classic” 7T scanner while subjects were watching a Human Connectome Project (HCP) movie. For layer-fMRI scanning without venous biases, a blood volume sensitive VASO (Lu 2003) sequence was used. We used the MAGEC VASO approach to maintain the VASO T1-weighting across long echo trains (Huber 2020).Scanning:
This study is based on prior experiments (shown in (Mueller 2021)). In order to find an improved sequence/reconstruction framework for the current study, 9 two-hour sessions in eight participants were performed (shown in Fig. 1-2). These newly improved scan/reconstruction parameters were then used for 8 two-hour sessions in the main single participant. These data are openly accessible on OpenNeuro (Markiewicz 2021): https://openneuro.org/datasets/ds003216. 5 more two-hour sessions datasets will be available soon. The final parameters that we used are: resolution = 0.842mm iso, TR = 5.1s (alternating 5.1s/5.2s), 3D-EPI (Poser 2010), GRAPPA 3x2, two segments and 3D-CAIPI 1 (equivalent to GRAPPA 6x1 with pre-phased CAIPI 3) (Poser 2013, Stirnberg 2021), TA = 14 min, 5 runs per session, the last two TRs were NORDIC (Vizioli 2021) noise scans (not included in reconstruction so far). This sequence is available via SIEMENS’ C2P ‘app store’ Teamplay.
Further acquisition parameters: https://layerfmri.page.link/WBprotocol. Respiratory and cardiac traces were recorded for vascular reactivity analyses.
Data processing:
Motion correction (ANTS, Avants 2008), BOLD correction in LayNii (Huber 2021), and segmentation of GM and WM were initially performed in FreeSurfer and then manually corrected.
Layer-connectivity analysis:
To investigate connectivity networks across layers, we performed ICA on run-averaged functional time series independently on three layer groups. For the columns of specific layer-profiles, we defined columns (LayNii LN2_COLUMNS), calculated ‘hubness’ (AFNI 3dTcorrMap) and performed PCA (AFNI 3dpc). Then, we highlighted individual layer-profiles and presented their columnar distribution across the brain.
Results and Discussion
Fig. 1 shows the newly incorporated and tested sequence advancements compared to previous work. The reduction of spatial and temporal artifacts with segmentation and shorter TRs are clearly visible.Fig. 2 depicts results from pilot experiments to find the best acceleration scheme (the winner was six-fold acceleration).
Fig. 3 depicts the preprocessing quality (alignment to anatomy, layerification in EPI, and alignment across runs).
Fig. 4 and Fig. 5 exemplify the neuroscientific applicability of the provided data. Fig. 4 depicts the layer-dependent differences of common ICA networks. While the Parieto-Frontal network, the Visual network, and the Auditory network show consistent topographical distributions across depths, the ‘default mode network’ is interestingly showing considerable deviations. We find that the visual and auditory networks show stronger z-scores in superficial and deep layers, compared to middle layers (Fig. 4). This finding might be related to the fact that neurons even in those uni-modal areas have more feedback synapses from cortical areas compared to feedforward input. When analyzing each session’s data separately, we see consistent layer signatures, demonstrating high reliability (Fig. 5A).
We used PCA analyses across all layer profiles in all columns to explore data-driven approaches of consistent layer profiles across areas. Doing so for movie watching tasks, we find that columns that exhibit layer-profiles of inverted U-shapes (feedforward-like) tend to be stronger represented in the parieto-frontal area. Whereas columns with U-shaped (feedback-like) layer-profiles tend to be strongly represented in visual areas.
Summary and Conclusion
At last year's ISMRM, we presented an attempt to acquire a whole-brain layer-specific connectivity protocol, with mixed success. This year, we went through a second iteration of re-considering all acquisition and reconstruction choices to reduce the TR, improve the artifact level, and provide another new and improved dataset of 25 runs of whole-brain layer-specific movie watching.The open dataset provided here, will be useful for benchmarking developing laminar preprocessing strategies. Furthermore, with the large number of runs provided of the same 14 min movie clips, this dataset will be helpful for a comprehensive characterization of the reproducibility layer-fMRI connectivity results.
Acknowledgements
Help with scanning:
These data were acquired with the kind support of scannexus at their 7T MAGENTOM “Classic” SIEMENS scanner. We thank Mathilde Kennis for assistance with after hour scans.
Funding:
Kenshu Koiso is funded by Japan Public-Private Partnership Student Study Abroad Program (TOBITATE! Young Ambassador Program), by Japan Student Services Organization (JASSO) and by UEC fund.
Laurentius Huber was funded by the NWO VENI project 016.Veni.198.032.
Benedikt Poser is partially funded by the NWO VIDI grant 16.Vidi.178.052, by the National Institute for Health grant R01MH/111444 (PI David Feinberg) and by the H2020 FET-Open AROMA grant agreement no. 88587.
Yoichi Miyawaki is funded by JSPS KAKENHI (20H00600, 18KK0311).
Ethics:
The scanning procedures have been approved by the Ethics Review Committee for Psychology and Neuroscience (ERCPN) at Maastricht University, following the principles expressed in the Declaration of Helsinki.
References
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26-41. doi:10.1016/j.media.2007.06.004
- Guidi M, Huber L, Lampe L, Merola A, Ihle K, Möller HE. Cortical laminar resting-state fluctuations scale with the hypercapnic bold response. HBM. 2020;41:2014-2027. doi:10.1002/hbm.24926
- Havlicek M, Uludag K. A dynamical model of the laminar BOLD response. Neuroimage. 2019;204:116209. doi:10.1101/609099
- Huber L, Finn ES, Chai Y, et al. Layer-dependent functional connectivity methods. Prog Neurobiol. 2020:in print. Doi:j.pneurobio.2020.101835
- Huber L (Renzo), Poser BA, Bandettini PA, et al. LayNii: A software suite for layer-fMRI. Neuroimage. 2021;237:118091. doi:10.1016/j.neuroimage.2021.118091
- Lu H, Golay X, Pekar JJ, van Zijl PCM. Functional magnetic resonance imaging based on changes in vascular space occupancy. Magn Reson Med. 2003;50:263-274. doi:10.1002/mrm.10519
- Markiewicz CJ, Gorgolewski KJ, Feingold F, et al. OpenNeuro: An open resource for sharing of neuroimaging data. Elife. 2021:10:e71774. doi: https://doi.org/10.7554/eLife.71774
- Mueller et al, 2021, ISMRM #0630
- Poser BA, Koopmans PJ, Witzel T, Wald LL, Barth M. Three dimensional echo-planar imaging at 7 tesla. Neuroimage. 2010;51(1):261-266. doi:10.1016/j.neuroimage.2010.01.108
- Poser BA, Ivanov D, Kemper VG, Kannengiesser SA, Uludag K, Barth M. CAIPIRINHA-accelerated 3D EPI for high temporal and / or spatial resolution EPI acquisitions. Esmrmb. 2013:226
- Pais-Roldán P, Yun SD, Palomero-Gallagher N, Shah NJ. Cortical depth-dependent human fMRI of resting-state networks using EPIK. bioRxiv. 2020:1-26. doi:10.1101/2020.12.07.414144
- Stirnberg R, Stöcker T. Segmented K-Space Blipped-Controlled Aliasing in Parallel Imaging (Skipped-CAIPI) for High Spatiotemporal Resolution Echo Planar Imaging. Magn Reson Med. 2020;85(0):1540-1551. doi:10.1101/2020.06.08.140699
- Shadlen MN, Kiani R. Decision making as a window on cognition. Neuron. 2013;80(3):791-806. doi:10.1016/j.neuron.2013.10.047
- Vizioli L, Moeller S, Dowdle L, et al. Lowering the thermal noise barrier in functional brain mapping with magnetic resonance imaging. Nat Commun. 2021;12:5181. doi:10.1038/s41467-021-25431-8
Figures
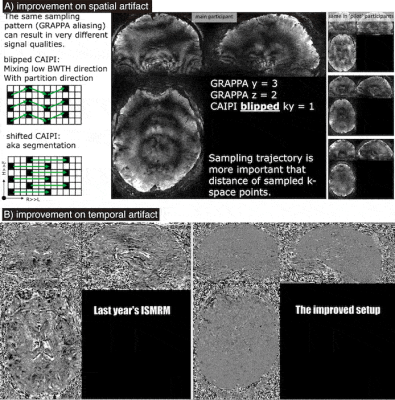
Panel A: Segmentation and kz-CAIPI can shorten TRs without ghosting artifacts that are common when mixing the low bandwidth direction with the partition direction. This insight allowed us to reduce the volume TR from 8.9 s to 5.1 s.
Panel B: Temporal artifact level of the improved sequence/reconstruction approach. This figure presents the temporal VASO signal evolution (demeaned) of movie watching data averaged across 20 runs (across 4 days).
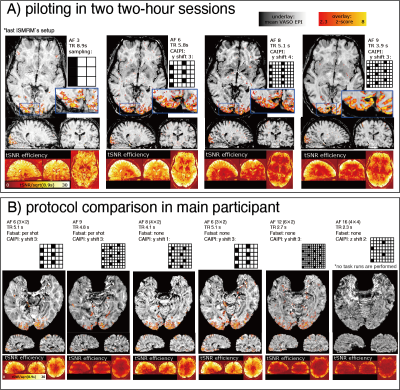
Fig2: Finding the best sampling approach
Panel A: Piloting before scanning the main participant. The middle left sequence (AF = 6, TR = 5.8 s, CAIPI y shift 3) has an advantageous tSNR efficiency.
Panel B: Protocol comparison in the main participant. The third column (AF = 6, TR = 5.1 s without fat saturation) depicts a good compromise of TR and artifact level. We decided to use this sequence for all subsequent scans.
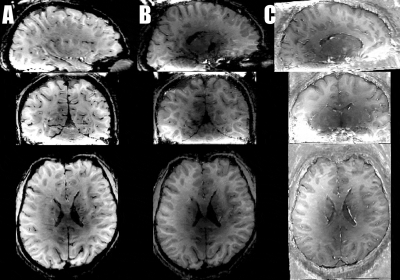
Fig3: Preprocessing quality (GIF animation)
Panel A: Non-linear alignment quality of a 3T MPRAGE to VASO EPI. Most of the brain is of usable quality.
Panel B: Layerification quality in the EPI space.
Panel C: Session-to-session alignment quality. The gif animation cycles across 5 separate sessions of multiple days.
Note that the spatially heterogeneous brightness level of the T1-weighted EPI is a feature of variable flip angles in the MAGEC-VASO approach and does not affect the functional contrast.
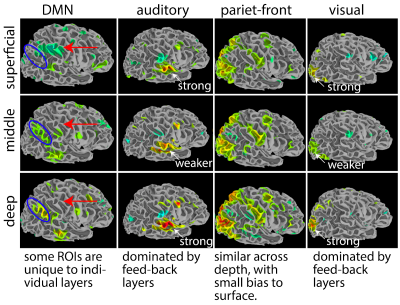
Fig4: Layer-dependent differentiation of ICA-determined connectivity networks
We find that the primary areas (e.g., visual and auditory) are showing stronger z-scores for superficial and deep layers. The DMN seems to subdivide into different sub-ROIs for different layers. Other networks (e.g., parietal-frontal) are relatively consistent across depths with minimal layer-differentiation. Note that the purpose of this figure is to exemplify the neuroscientific usability of the provided data.
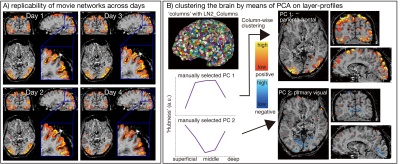
Fig5: Network detection replicability and layer-profile analysis
Panel A: GLM results of a representative brain network shown independently for each day. We can see consistent layer signatures for each day's result.
Panel B: PCA extracted columns with feedforward-like and feedback-like layer-profiles. The columns (top left in B) are generated with the LayNii command “LN2_Columns”. The lower left graphs are the two representative layer-profile PCs. The right figures in panel B are the columns with the selected layer-profile PCs accordingly.