0200
A Deep-Learning Approach to Predicting Disease Progression in Multiple Sclerosis Using Magnetic Resonance Imaging
Loredana Storelli1, Matteo Azzimonti1,2, Mor Gueye1,2, Paolo Preziosa1,2, Carmen Vizzino1, Gioacchino Tedeschi3, Nicola De Stefano4, Patrizia Pantano5,6, Massimo Filippi1,2,7,8,9, and Maria A. Rocca1,2,9
1Neuroimaging Research Unit, Division of Neuroscience, IRCCS San Raffaele Scientific Institute, Milan, Italy, 2Neurology Unit, IRCCS San Raffaele Scientific Institute, Milan, Italy, 3Department of Advanced Medical and Surgical Sciences, and 3T MRI-Center, University of Campania “Luigi Vanvitelli”, Maples, Italy, 4Department of Medicine, Surgery and Neuroscience, University of Siena, Siena, Italy, 5Department of Human Neurosciences, Sapienza University of Rome, Rome, Italy, 6IRCCS NEUROMED, Pozzilli, Italy, 7Neurorehabilitation Unit, IRCCS San Raffaele Scientific Institute, Milan, Italy, 8Neurophysiology Service, IRCCS San Raffaele Scientific Institute, Milan, Italy, 9Vita-Salute San Raffaele University, Milan, Italy
1Neuroimaging Research Unit, Division of Neuroscience, IRCCS San Raffaele Scientific Institute, Milan, Italy, 2Neurology Unit, IRCCS San Raffaele Scientific Institute, Milan, Italy, 3Department of Advanced Medical and Surgical Sciences, and 3T MRI-Center, University of Campania “Luigi Vanvitelli”, Maples, Italy, 4Department of Medicine, Surgery and Neuroscience, University of Siena, Siena, Italy, 5Department of Human Neurosciences, Sapienza University of Rome, Rome, Italy, 6IRCCS NEUROMED, Pozzilli, Italy, 7Neurorehabilitation Unit, IRCCS San Raffaele Scientific Institute, Milan, Italy, 8Neurophysiology Service, IRCCS San Raffaele Scientific Institute, Milan, Italy, 9Vita-Salute San Raffaele University, Milan, Italy
Synopsis
Artificial intelligence (AI) approaches have been applied in several fields of multiple sclerosis (MS) in recent years. However, their application to predict disease progression remains largely unexplored. In this study, we obtained a robust and accurate AI tool for predicting clinical and cognitive evolution at two years for MS patients, based on just T1-weighted and T2-weighted brain MRI scans at baseline visit, which exceeded the performance of two expert physicians blinded to patients’ clinical history. This algorithm has the potential to be an important tool to support physicians for a prompt recognition of MS patients at risk of disease worsening.
Introduction
Magnetic resonance imaging (MRI) is an important in vivo tool for diagnosis and monitoring disease course and treatment in multiple sclerosis (MS).1 However, the prognostic value of MRI for predicting disease evolution in these patients is still debated. The possibility to predict disease progression in MS before the accumulation of irreversible clinical disability would be very important for promptly managing personalized treatment, but it remains an unmet need.2 Artificial intelligence and, in particular, deep-learning approaches have rapidly become popular mathematical models to make predictions without human intervention.3 In the field of MS, the application of deep-learning algorithms to predict disease progression remains largely unexplored. Thus, the aim of this study was to develop and apply a deep-learning algorithm on a large multicenter cohort of patients collected from the Italian Neuroimaging Network Initiative (INNI) to predict disease evolution (based on clinical disability and cognitive impairment) at two years of follow-up in MS patients from their baseline MRI features. The performance of the algorithm was finally compared to that of two expert physicians.Methods
For 373 MS patients, baseline T2-weighted and T1-weighted brain MRI 3T scans, as well as baseline and two-year clinical and cognitive assessments were collected from the INNI repository. The collected images were quality controlled and underwent a common pre-processing including: lesion-filling, segmentation of brain tissues and an affine coregistration in the MNI atlas space to obtain anatomical comparable brain structures among the patients. 325 patients from the four INNI promoter Centers were used to train and optimize the algorithm, while 48 patients from just one Center but with the MRI acquired on a different scanner, in respect to the training set, were used as an independent test set. A deep-learning architecture based on 3 convolutional neural networks (CNN) blocks for each MRI modality, finally concatenated, was implemented (Figure 1) to predict: (1) clinical worsening (Expanded Disability Status Scale [EDSS]-based model), (2) cognitive deterioration (Symbol Digit Modalities Test [SDMT]-based model), or (3) both (EDSS+SDMT-based model). In parallel, two expert physicians, blinded to the identity and clinical history of each patient, performed a visual assessment of the baseline MRI dataset. They independently classified patients as having a negative or positive prognosis according to five MRI criteria: (1) at least nine T2-hyperintense brain lesions; (2) at least one T2-hyperintense infratentorial lesion; (3) at least one T1-hypointense cortical/juxtacortical lesion; (4) brain atrophy, evaluated qualitatively; (5) at least nine T1-hypointense brain lesions. The presence of at least 3 of these criteria was considered indicative of a negative clinical prognosis. The accuracy, sensitivity and specificity of the method was tested and compared to the performance of the two expert physicians.Results
At follow-up, 97 MS patients had worsened clinically and fifteen relapsing-remitting MS patients evolved to secondary progressive MS. 38 MS patients showed cognitive worsening after two years and 14 patients showed both clinical and cognitive worsening. After optimizing the model on the training set, we found an accuracy of 83.3% for the prognosis of clinical disability worsening at follow-up on the independent test set, with 42.9% of sensitivity and 93.2% of specificity. For the prediction of cognitive worsening (SDMT-based), the CNN model achieved an accuracy of 67.7% in the test set, with a sensitivity of 60% and a specificity of 81.8%. In Figures 2 and 3, two example of correctly and wrongly classified patients by the algorithm are provided. When combining clinical and cognitive information to train the model (EDSS+SDMT-based), the deep-learning algorithm reached 85.7% accuracy, 96% sensitivity and 75% specificity. With this trained model, 75% of worsened patients were identified by the algorithm, while 87.5% of clinically and cognitively stable patients were correctly predicted. Considering the test set only, expert raters showed an accuracy of 70% for correct disease prognosis, with a sensitivity of 15.1% and a specificity of 80%. However, for this dataset, only 14.3% of worsened EDSS patients were correctly identified as disease worsened, while 80% of the stable patients were correctly recognized.Discussion
The deep-learning algorithm trained to predict clinical evolution using EDSS information associated with MRI showed higher performance in the test-set, with higher accuracy, sensitivity and specificity compared to the human experts. This algorithm has the potential to be an important tool for supporting, rather than replacing, physicians in their clinical routine for the prompt management of MS patients at risk of disease worsening. Artificial intelligence could be an aid to clinicians in the difficult task of handling large amounts of data and understanding it (i.e., extracting important features and patterns), while conversely, the knowledge and expertise of clinicians would improve artificial intelligence performance. In future, the capabilities of radiologists may be improved and broadened by these tools.Conclusions
We developed a robust and accurate model for predicting clinical and cognitive worsening of MS patients after two years, based on conventional T2-weighted and T1-weighted brain MRI scans obtained at baseline. This algorithm may be valuable for supporting physicians in their clinical practice for the earlier identification of MS patients at risk of disease worsening.Acknowledgements
This study was partially supported by Fondazione Italiana Sclerosi Multipla with a research fellowship (FISM 2019/BR/009) and research grants (FISM2018/R/16; FISM2018/S/3), and financed or co-financed with the ‘5 per mille’ public funding.References
1. Filippi M, Rocca MA. MR imaging of multiple sclerosis. Radiology 2011;259:659-681. 2. Filippi M PP, Langdon D, Lassmann H, Paul F, Rovira A, Schoonheim MM, Solari A, Stankoff B, Rocca MA. Identifying Progression in Multiple Sclerosis: New Perspectives. Ann Neurol 2020;88(3). 3. Bernal J, Kushibar K, Asfaw DS, et al. Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: a review. Artif Intell Med 2019;95:64-81.Figures
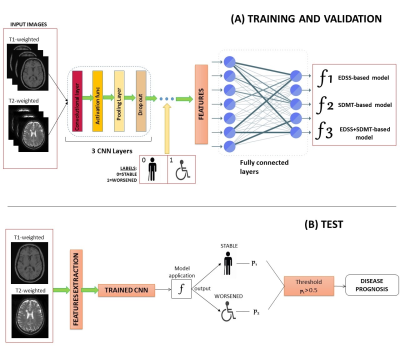
Fig. 1 A schematic overview of the deep-learning
network architecture implemented to train and optimize the model (in A), and to
finally test it on an independent dataset (in B).
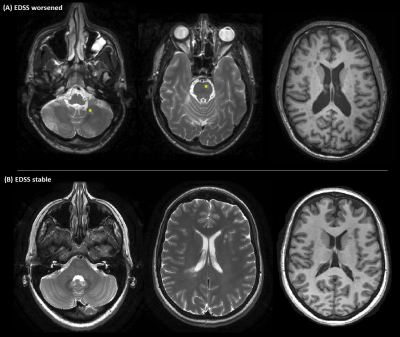
Fig. 2 Two patients correctly classified by the deep-learning
algorithm are shown. A) a clinically worsened patient at follow-up (according
to EDSS score change) showing T2-hyperintense infratentorial lesions (yellow
stars) and evident brain atrophy at baseline. B) a clinically stable patient at
follow-up. In contrast to patient in A, he showed absence of T2-hyperintense
infratentorial lesions and little brain atrophy at baseline. First two columns:
axial T2-weighted brain MRI; last column: axial T1-weighted brain MRI.
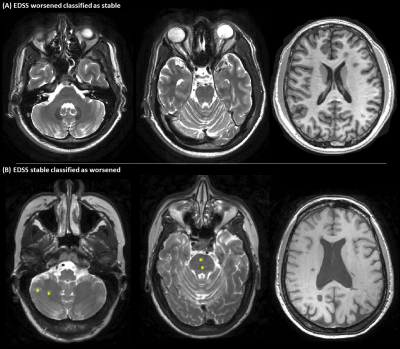
Fig. 3 Two patients wrongly classified by the
deep-learning algorithm. A) clinically worsened patient at follow-up (according
to EDSS score change) classified as stable by the algorithm. The patient showed
a moderate T2-hyperintense lesion load, and did not present infratentorial
lesions or evident brain atrophy at baseline. B) clinically stable patient
classified as EDSS worsened by the algorithm. The patient showed infratentorial
lesions (yellow stars) and pronounced brain atrophy. First two columns: axial
T2-weighted brain MRI; last column: axial T1-weighted brain MRI.
DOI: https://doi.org/10.58530/2022/0200