0175
ViT-PU-Net: Volumetric Phase Unwrapping for MR images based on Vision Transformer1Electrical and Electronic Engineering, Yonsei University, Seoul, Korea, Republic of, 2Radiology, Stanford University, Stanford, CA, United States
Synopsis
MR phase information has been used in many MRI applications, recently, so it is very important to estimate the correct MR phase. Since the MR phase is encoded in complex exponential, the actual phase information obtained is in the form of a wrapped signal. Unfortunately, conventional MR phase unwrap methods are highly related to SNR and require a lot of computation, so an efficient method is needed.. Here, we propose ViT-PU-Net to perform 3D phase unwrap using ViT-based encoder architecture. Therefore, we investigated whether this architecture could be applied to 3D phase unwrap task by applying it to various data.
Introduction
In recently, MR phase information has been used in many MRI applications such as correcting image distortion, Quantification of iron-rich contents and temperature changes in tissue1-2. However, since accurate phase unwrapping estimation requires high computational complexity, an efficient method has been required. Specifically, the PRELUDE method, which is classified as the golden standard in MRI, is the most robust phase unwrapping method, but has recently been replaced by a more efficient algorithm such as SEGUE3-4. Unfortunately, conventional methods did not overcome the trade-off in performance, and recently, single-step phase estimation by applying deep-learning has been studied5-6. The point is that most of the studies select CNN as the baseline model, but it was confirmed that the locality of CNN was not suitable for the phase unwrapping task9. Therefore, we propose ViT-PU-Net, the first study to perform 3D phase unwrapping inspired by Vision transformer that can focus on global features. Vision Transformer has recently applied a Transformer-based Encoder in the medical imaging field, showing potential in the field of handling volumetric data such as 3D segmentation and registration, and has the potential to show that it is an architecture suitable for the 3D phase unwrapping work11-12.Methods
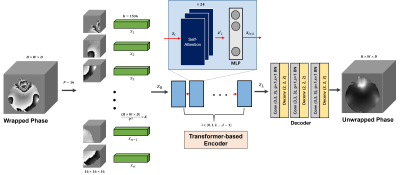
[Network architecture]In Figure 1, the network structure consists of three parts, and the first part is patch embedding. A training instance as input is created to a 3D patch (P=16) and linear projection is performed. Here, the flattened vector { x1, x1 ... xn} are embedded z0 and where n=(HxWxD) / P3. The dimension K=1536 of the flattened vector used at this time was determined empirically. The second step, the ViT-based encoder part, encodes with L=12 stacked structure of multi-head self-attention (MSA) layers. In addition, the dimension of the MLP layer of the Transformer block was selected as 3096 to secure the capability. The last decoder is a stacked structure of the up-sampler of the CNN backbone in a cascade form, and the parameters used can be referred to the figure.
[Training]
We use 3D mGRE data with 8-echos from 28 healthy in-vivo data to train the network. Of the total 216 instances, 184 were divided into the train set, 24 as the validation set, and 8 as the test set. All datasets consist of wrapped phase and PRELUDE pairs, and brain mask extraction13 and prelude using FSL tool15 were used in the unwrapping process with volume size [240, 240, 64]. For the network training, data augmentation such as random filp and affine transforms are used, and weighted variance loss is used for the loss function9.
L(output, Y) = E[ M×(output - Y)2 ] / E [M] - E[ (M×(output - Y)) / E [M] ]2, where Y denotes PRELUDE, M is Magnitude of a MR image.
[Testing]
First, we evaluate the result of the test set and observe the signal variation of one slice and compare it with the phase output of ViT-PU-Net. Next, SEGUE and PRELUDE are compared for the QSM challenge 2016 data to test the generalization of the network.
Results
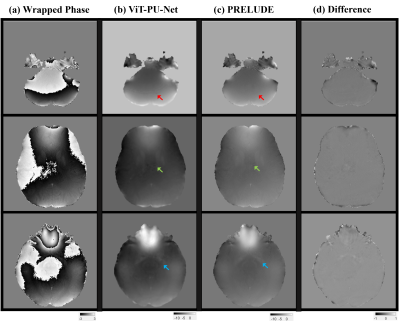
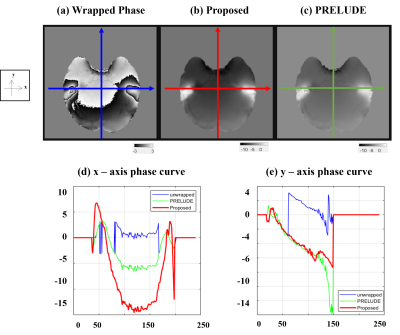
Figure 2 shows the results for the test set, and it can be seen that the ViT-based network is well trained. Overall, compared to (b) PRELUDE, (a) proposed can be seen that there is a tendency to under-estimation, and blurring can also be observed. And in the difference (c), the residual is observed at the boundary where the wrap jump occurs.Figure 3 shows the results for also test set, which observed the signal variation on the x and y axes for a specific slice. As can be seen from the graph, it can be observed that the x-axis results vary considerably in the proposed method. On the other hand, it can be seen that the y-axis results follow the trend to some extent. It can be seen that the prediction is lowered for the x-axis with a lot of phase jumping.
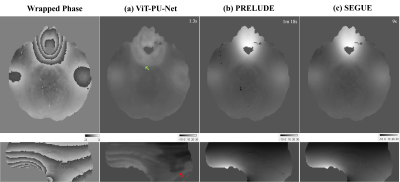
In figure 4, it is the result of QSM challenge data14 taken in a different scanner, and compared to the conventional method, it is vulnerable to severe phase variation.
Discussion and Conclusion
It has been recently proven in several studies that the Vision transformer-based encoder has the advantage of being able to focus on global features. This encoding method has the potential to learn contextual information, especially for large 3D data unlike existing CNNs. In this study, the pure ViT encoder used in the proposed architecture shows a lot of blurring and eventually loses phase information. In addition, 3T also suffered from generalization problems for data obtained from other scanners, and the more severe the phase variation, the more the network lost information. Therefore, in order to learn the severe wrap count while preserving this detailed information, a more delicate technique such as ViT-CNN seems to be needed.Acknowledgements
No acknowledgement found.References
[1]. ROBINSON, Simon Daniel, et al. An illustrated comparison of processing methods for MR phase imaging and QSM: combining array coil signals and phase unwrapping. NMR in Biomedicine, 2017, 30.4: e3601.
[2] CHAVEZ, Sofia; XIANG, Qing-San; AN, Li. Understanding phase maps in MRI: a new cutline phase unwrapping method. IEEE transactions on medical imaging, 2002, 21.8: 966-977.
[3] JENKINSON, Mark. Fast, automated, N‐dimensional phase‐unwrapping algorithm. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine, 2003, 49.1: 193-197.
[4] KARSA, Anita; SHMUELI, Karin. SEGUE: A speedy region-growing algorithm for unwrapping estimated phase. IEEE transactions on medical imaging, 2018, 38.6: 1347-1357.
[5] SPOORTHI, G. E.; GORTHI, Subrahmanyam; GORTHI, Rama Krishna Sai Subrahmanyam. PhaseNet: A deep convolutional neural network for two-dimensional phase unwrapping. IEEE Signal Processing Letters, 2018, 26.1: 54-58.
[6] ZHOU, Hongyu, et al. The PHU‐NET: A robust phase unwrapping method for MRI based on deep learning. Magnetic Resonance in Medicine, 2021.
[7] YANG, Fangshu, et al. Robust phase unwrapping via deep image prior for quantitative phase imaging. IEEE Transactions on Image Processing, 2021, 30: 7025-7037.
[8] RONNEBERGER, Olaf; FISCHER, Philipp; BROX, Thomas. U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015. p. 234-241.
[9] RYU, Kanghyun, et al. Development of a deep learning method for phase unwrapping MR images. In: Proc Intl Soc Mag Reson Med. 2019. p. 4707.
[10] DOSOVITSKIY, Alexey, et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929, 2020.
[11] HATAMIZADEH, Ali, et al. Unetr: Transformers for 3d medical image segmentation. arXiv preprint arXiv:2103.10504, 2021.
[12] CHEN, Junyu, et al. ViT-V-Net: Vision Transformer for Unsupervised Volumetric Medical Image Registration. arXiv preprint arXiv:2104.06468, 2021.
[13] SMITH, Stephen M. Fast robust automated brain extraction. Human brain mapping, 2002, 17.3: 143-155.
[14] LANGKAMMER, Christian, et al. Quantitative susceptibility mapping: Report from the 2016 reconstruction challenge. Magnetic resonance in medicine, 2018, 79.3: 1661-1673.
[15] JENKINSON, Mark, et al. Fsl. Neuroimage, 2012, 62.2: 782-790.
Figures