0173
Multi-scale UNet with Self-Constructing Graph Latent for Deformable Image Registration1Department of Biomedical Magnetic Resonance, Otto von Guericke University Magdeburg, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University Magdeburg, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University Magdeburg, Magdeburg, Germany, 4German Centre for NeurodegenerativeDiseases, Magdeburg, Germany, 5Center for Behavioral Brain Sciences, Magdeburg, Germany, 6Leibniz Institute for Neurobiology, Magdeburg, Germany
Synopsis
Deep Learning based deformable registration techniques such as Voxelmorph, ICNet, FIRE, do not explicitly encode global dependencies and track large deformations. This research attempts to encode semantics, i.e. structure and overall view of the anatomy in the supplied image, by incorporating self-constructing graph network in the latent space of a UNet model. It also attempts to track larger deformations through multiscale architecture and maintains consistent deformations through cycle consistency. The proposed method was compared against Voxelmorph and ANTs for T1 intramodal and T1-T2 Intermodal registration on IXI Dataset. The experiments show that the proposed model outperforms the baselines.
Introduction
Deep Learning-based techniques have been applied successfully to tackle various medical image processing problems. Over the years, several image registration techniques have been proposed using deep learning. Deformable image registration techniques such as Voxelmorph1 have been successful in capturing finer changes and providing smoother deformations. Voxelmorph1 is a deep learning based deformable image registration method, using a UNet-like model that takes in affinely aligned fixed and moving images and performs deformable registration. However, Voxelmorph1, as well as ICNet2 and FIRE3 do not explicitly encode global dependencies (i.e. overall anatomical view of the supplied image) and track large deformations. This research improves upon Voxelmorph1 by employing self-constructing graph network (SCGNet)4 to encode semantics – which can improve the learning process of the model and help the model to generalise better, multi-scale supervision – to be able to work well in case of small as well as large deformations, and cycle consistency – for making the deformations consistent.Methods
The proposed method tries to capture global dependencies using SCGNet4, wherein encoded CNN features are used to find out semantics. The authors hypothesise that these dependencies could help the model to learn relationships even between two distantly located structures in the fixed and moving image and help generate better deformations. Multi-scale supervision was employed to handle different amounts of deformations, where deformations are predicted, and losses are calculated for both the original image and a downsampled image. The authors hypothesised that deformations on downsampled images would cover for larger ones while the deformations on the original image would produce finer ones. Regularisation of sharp deformation was also used to obtain smooth deformation fields. An adapted version of the cycle consistency loss, inspired by ICNet2, was employed to maintain consistent deformations. The proposed method was compared against and Voxelmorph1 and non-deep learning-based deformable registration using Symmetric Normalisation (SyN) of the ANTs5.Model Description
The model uses a UNet-like structure to encode-decode the image information. For encoding, it uses convolution with stride to downsample the image. The encoded information is fed into the latent space of the SCGNet4, which first parametrises the encodings to latent embeddings with mean and standard deviation, and derives the undirected graph through an inner product of latent embeddings. The output is then fed into a fully-connected graph convolution network (GCN) which attempts to learn the semantics of the overall anatomy. The GCN output is upsampled through interpolation and supplied to a CNN decoder which takes in also input from the respective encoder as skip connections. Finally, deformation fields are smoothened by convolution layers. Fig. 1. illustrates the model architecture.
Training Procedure
Fixed (F) and moving (M) images were first downsampled with a factor of 2 in all dimensions, then they were concatenated and sent as an input to the network. Network processed the images and provided deformation fields at both original and a downsampled scale. They were then applied on both original, downsampled moving images and compared with the corresponding fixed images with a set of loss functions. For cycle consistency loss, this process is repeated for the same image pair by swapping the fixed and moving images. Finally, both pairs of losses are added up and backpropagated through the network. All the loss functions used can be defined with the following equation:
$$Loss=-1.2*(SIM\_original_{F->M}+SIM\_original_{M->F})\\-0.6*(SIM\_downsampled_{F->M}+SIM\_downsampled_{M->F})\\+0.5*(ST\_original_{F->M}+ST\_original_{M->F})\\+0.25*(ST\_downsampled_{F->M}+ST\_downsampled_{M->F})\\+5.0*(SCG_{F->M}+SCG_{M->F})$$
where $$$SIM$$$ denotes similarity measure - which was normalised cross-correlation and normalised mutual information for intramodal and intermodal registrations respectively, $$$ST$$$ is the smoothness loss, $$$SCG$$$ denotes the SCG loss, $$$\_original$$$ signifies the volumes in the original scale, and $$$\_downsampled$$$ signifies the downsampled version of the volumes. This loss was minimised using the Adam optimiser with a learning rate of 0.0001 for 700 epochs.
Data
T1w and T2w images from the publicly available IXI dataset6 were used in this research. Before training the proposed network, the images were pre-processed using FreeSurfer7 in a similar manner as Voxelmorph1. 200 volumes were used during training and 50 for validation. T1w intramodal registrations were performed between different subjects, and intermodal registrations with T1w as fixed and T2w as moving were performed using the T1-T2 pairs of same subjects.
Results
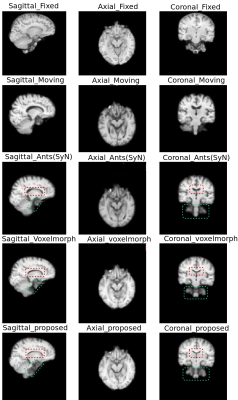
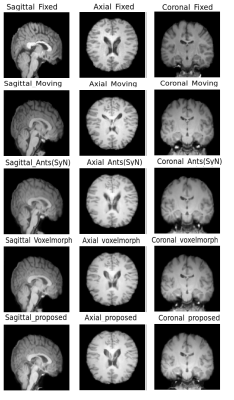
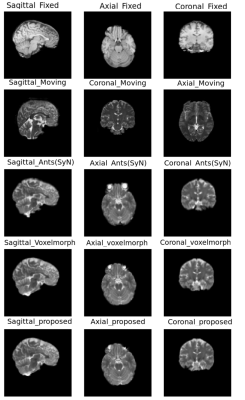
The quantitative evaluation was performed using SSIM8 and Dice scores for white matter, grey matter and CSF, between fixed and registered images. The proposed model performs better than the baselines, with a mean SSIM of 0.9808 and a mean Dice score of 0.8013 for intramodal registration. The results were found to be statistically significant against the baselines after evaluating with two-tailed t-tests. For intermodal registration, even though the mean Dice score of the proposed method was better than the baselines, they were not statistically significant. Table.1 shows the quantitative evaluation. The proposed network also generalises better than Voxelmorph, as demonstrated in Table.2, while testing on images that were not used in training and even when they were pre-processed differently. Fig 3-5 show the results qualitative results of the different methods.Conclusion
The experiments showed that the prosed network outperformed the baselines with statistical significance for intramodal registration, quantified using SSIM and Dice scores. For intermodal registration, improvements were observed both qualitatively and quantitatively. However, the quantitative improvements over the baselines were not statistically significant.Acknowledgements
This work was in part conducted within the context of the International Graduate School MEMoRIAL at OvGU (Project no. ZS/2016/08/80646).References
[1] Balakrishnan, Guha, et al. "Voxelmorph: a learning framework for deformable medical image registration." IEEE transactions on medical imaging 38.8 (2019): 1788-1800.
[2] Zhang, Jun. "Inverse-consistent deep networks for unsupervised deformable image registration." arXiv preprint arXiv:1809.03443 (2018).
[3] Wang, Chengjia, et al. "FIRE: unsupervised bi-directional inter-modality registration using deep networks." arXiv preprint arXiv:1907.05062 (2019).
[4] Liu, Qinghui, Michael Kampffmeyer, and Robert Jenssen. "Self-constructing graph convolutional networks for semantic labeling." IGARSS 2020-2020 IEEE International Geoscience and Remote Sensing Symposium. IEEE, 2020.
[5] Avants, Brian B., Nick Tustison, and Gang Song. "Advanced normalization tools (ANTS)." Insight j 2.365 (2009): 1-35. Python implementation of ANTs URL: https://github.com/ANTsX/ANTsPy
[6] IXI Dataset by brain-development.org. URL: https://brain-development.org/ixi-dataset/
[7] Fischl, Bruce. "FreeSurfer." Neuroimage 62.2 (2012): 774-781.
[8] Wang, Zhou, et al. "Image quality assessment: from error visibility to structural similarity." IEEE Transactions on Image Processing, vol. 13, no. 4 (2004): 600-612.
Figures
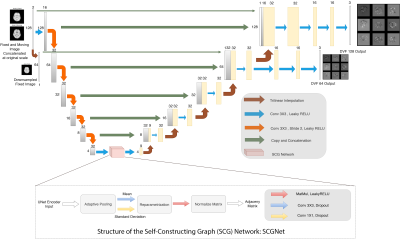
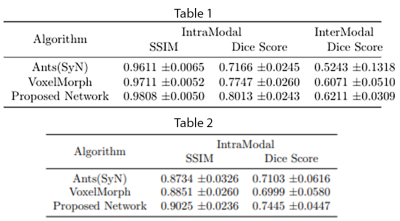
Table 1: Quantitative analysis averaging SSIM and Dice scores over 50 test subjects for Ants Symmetric Normalization (SyN), Voxelmorph and the proposed network on the train and test images pre-processed using the same pre-processing pipeline - for intra-modal (T1w) and inter-modal (T1w-T2w) registration.Table 2: Quantitative analysis over 30 test subjects for the three different methods on test images having a different pre-processing pipeline compared to images used to train the network - intra-modal (T1w) registration.