0172
StRegA: Unsupervised Anomaly Detection in Brain MRIs using Compact Context-encoding Variational Autoencoder1Department of Biomedical Magnetic Resonance, Otto von Guericke University Magdeburg, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University Magdeburg, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University Magdeburg, Magdeburg, Germany, 4MedDigit, Department of Neurology, Medical Faculty, University Hospital, Magdeburg, Germany, 5Center for Behavioral Brain Sciences, Magdeburg, Germany, 6German Centre for NeurodegenerativeDiseases, Magdeburg, Germany, 7Leibniz Institute for Neurobiology, Magdeburg, Germany
Synopsis
Deep learning methods are typically trained in a supervised with annotated data for analysing medical images with the motivation of detecting pathologies. In the absence of manually annotated training data, unsupervised anomaly detection can be one of the possible solutions. This work proposes StRegA, an unsupervised anomaly detection pipeline based on a compact ceVAE and shows its applicability in detecting anomalies such as tumours in brain MRIs. The proposed pipeline achieved a Dice score of 0.642±0.101 while detecting tumours in T2w images of the BraTS dataset and 0.859±0.112 while detecting artificially induced anomalies.
Introduction
Expert interpretation of anatomical images of the human brain is the central part of neuro-radiology. Several machine learning-based techniques have been proposed in recent times to assist in the analysis process. However, typically the ML models need to be trained to analyse or perform a certain specific task: e.g., brain tumour segmentation and classification. Not only do the corresponding training data require laborious manual annotations, but a wide variety of abnormalities can be present in a human brain MRI, which is challenging to represent in a given dataset. Hence, a possible solution can be an unsupervised anomaly detection (UAD)1 system that can learn a data distribution from an unannotated dataset of healthy subjects and then be applied to detect out of distribution samples. Such a technique can then be used to detect pathologies, for example brain tumours, without explicitly training the model for a specific task. Many Variational Autoencoder (VAE) based techniques2,3 have been proposed in the past for this task, but many of them perform poorly while detecting pathologies well in clinical data. This research proposes a compact version of “context-encoding” VAE (ceVAE)3 model, combined with pre and post-processing steps, creating a UAD pipeline which is more robust on clinical dataMethods
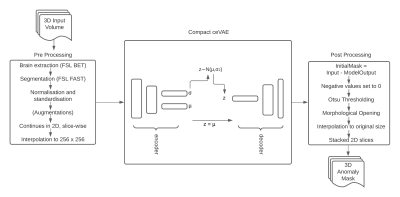
The proposed processing pipeline, StRegA: Segmentation Regularised Anomaly (Fig. 1), is based on a modified version of the ceVAE3 - Compact ceVAE (cceVAE), combined with pre and post-processing steps. Anomaly detection is performed in 2D, per-slice basis, and finally, the slices are stacked together to obtain the final result in 3D.The proposed cceVAE model is a modified and compact version of the ceVAE3 model. The core idea of ceVAE is to combine the reconstruction term with a density-based anomaly scoring. This allows better latent representation, which improves pixel and sample results3. The proposed cceVAE model includes a smaller encoder-decoder than the base ceVAE, with symmetric 64,128,256 feature maps and a latent variable size of 256, and also uses batch normalisation and residual connections. The output reconstruction of the model is then post-processed after subtracting it from the input to detect the anomaly.
The training was performed using only healthy subjects. Loss during training was calculated using a combination of Kullback–Leibler divergence and reconstruction loss using L1 loss - both with coefficients of 0.5, for the VAE part and similarly for the context encoding step. Combined losses were optimised using the Adam optimiser with a learning rate of 0.0001 for 60 epochs. During training, the network computes the latent representation of the non-anomalous input mean (μ) and standard deviation (σ) of the Gaussian distribution – consequently learning the feature distribution of the healthy dataset. During inference, only the mean of the distribution is used. Such a network, which has learnt the anomaly-free distribution, fails to reconstruct anomalies during inference.
The proposed method was trained using a combination of MOOD5 and IXI6 datasets; the testing was performed on the BraTS dataset7 and a held-out dataset from MOOD+IXI. This held-out dataset was used to test for anomaly-free and artificial-anomaly scenarios. For the second scenario, anomalies were added artificially to the held-out dataset with image processing. The MOOD dataset also came with a toy test dataset, which was used during testing as well. Two separate trainings were performed by using MOOD+IXI-T1 and MOOD+IXI-T2 datasets and were evaluated separately.
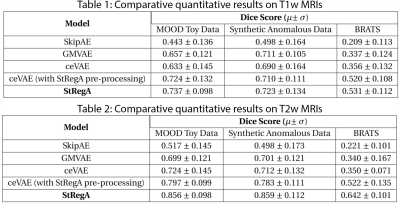
StRegA was compared against three baseline methods: SkipAE8, GMVAE9, and ceVAE3 using Dice scores, and the significance of differences was evaluated using independent t-tests. Additionally, one more comparison was performed against the ceVAE model by using the pre-processing techniques of StRegA.
Results
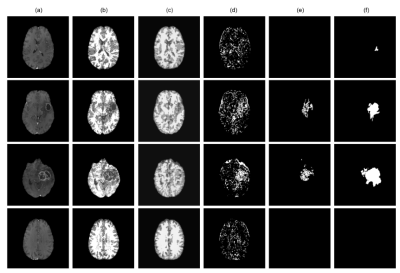
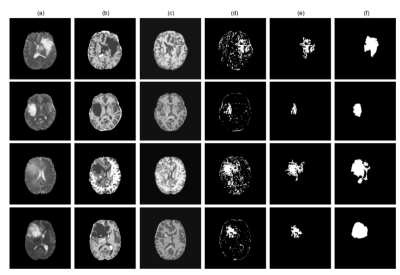
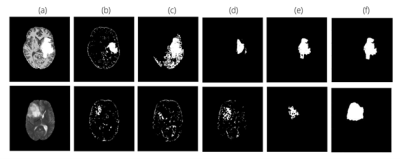
The proposed method achieved statistically significant improvement over all the baseline models while achieving 49% and 82% improvements in Dice scores over the baseline ceVAE3 while segmenting tumours from T1w and T2w brain MRIs from the BraTS dataset. The method performed better on the T2w images than the T1w ones. Fig. 2 and 3 show T1w and T2w results of StRegA from the BraTS dataset. Tables 1 and 2 show the quantitative results for T1w and T2w images, respectively. Fig. 4 shows a comparison of different methods while testing on T1w synthetic anomalous data and T2w MRI from the BraTS dataset.Discussion
Even though StRegA has performed significantly better than the baseline models, it is to be noted that the segmentations are not perfect, and there is a strong dependency on the pre-processing steps, as can be seen from the results of ceVAE and ceVAE with StrRegA pre-processing. Undersegmentations can be observed in all the examples. This also resulted in a complete disappearance of a small anomaly (e.g. Fig. 2 row 1). Nevertheless, these models were trained in an unsupervised fashion – without any labelled training data and showed its potential to be used as part of a decision support system.Conclusion
This work proposes an unsupervised anomaly detection pipeline, StRegA, using the proposed cceVAE model. The pipeline was trained and tested on T1w and T2w images and outperformed the baseline methods with statistical significance. StRegA achieved Dice scores of 0.531±0.112 and 0.642±0.101 while segmenting tumours, as well as 0.723±0.134 and 0.859±0.112 while segmenting artificial anomalies, from T1w and T2w MRIs, respectively.Acknowledgements
This work was in part conducted within the context of the International Graduate School MEMoRIAL at OvGU (project no. ZS/2016/08/80646) and supported by the federal state of Saxony-Anhalt (“I 88”).References
[1] Paul Bergmann, Michael Fauser, David Sattlegger, and Carsten Steger. MVTec AD – A Comprehensive Real-World Dataset for Unsupervised Anomaly Detection. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pages 9592–9600, 2019.
[2] Chatterjee, Soumick Sciarra, Alessandro & Dünnwald, Max & Agrawal, Shubham & Tummala, Pavan & Setlur, Disha & Kalra, Aman & Jauhari, Aishwarya & Oeltze-Jafra, Steffen & Speck, Oliver & Nürnberger, Andreas. Unsupervised reconstruction based anomaly detection using a Variational Auto Encoder. ISMRM 2021
[3] David Zimmerer, Simon AA Kohl, Jens Petersen, Fabian Isensee, and Klaus H Maier-Hein. Context-encoding variational autoencoder for unsupervised anomaly detection. arXiv preprint arXiv:1812.05941, 2018.
[4] Mark Jenkinson, Christian F Beckmann, Timothy EJ Behrens, Mark W Woolrich, and Stephen M Smith. Fsl. Neuroimage, 62(2):782–790, 2012.
[5] David Zimmerer et al. Medical Out-of-Distribution Analysis Challenge 2021. Zenodo. DOI: 10.5281/zenodo.4573948
[6] IXI Dataset https://brain-development.org/ixi-dataset/
[7] B. H. Menze, A. Jakab, S. Bauer, J. Kalpathy-Cramer, K. Farahani, J. Kirby, et al. "The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS)", IEEE Transactions on Medical Imaging 34(10), 1993-2024 (2015) DOI: 10.1109/TMI.2014.2377694
[8] Baur, Christoph and Wiestler, Benedikt and Albarqouni, Shadi and Navab, Nassir. “Bayesian Skip-Autoencoders for Unsupervised Hyperintense Anomaly Detection in High Resolution Brain Mri”, 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), 2020.
[9] Xiaoran Chen and Suhang You and Kerem Can Tezcan and Ender Konukoglu. “Unsupervised Lesion Detection via Image Restoration with a Normative Prior”, arXiv: 2005.00031.
Figures