0171
Learning to segment brain tumours using an explainable classifier1Department of Biomedical Magnetic Resonance, Otto von Guericke University Magdeburg, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University Magdeburg, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University Magdeburg, Magdeburg, Germany, 4Institute for Medical Engineerin, Otto von Guericke University Magdeburg, Magdeburg, Germany, 5Department of Biomedical Data Science, Stanford University, Stanford, CA, United States, 6Center for Behavioral Brain Sciences, Magdeburg, Germany, 7German Centre for NeurodegenerativeDiseases, Magdeburg, Germany, 8Leibniz Institute for Neurobiology, Magdeburg, Germany
Synopsis
Deep learning pipelines typically require manually annotated training data and the complex reasoning done by such methods make them appear as “black-boxes” to the end-users, leading to reduced trust. Unsupervised or weakly-supervised techniques could be a possible candidate for solving the first issue, while explainable classifiers or applying post-hoc interpretability-explainability methods on opaque classifiers may solve the second issue. This research addresses both problems by segmenting brain tumours without segmentation labels for training, using an explainable deep learning-based classifier. The classifier combined with a global pooling operation with a segmentation model to train and obtain classification results from this method.
Introduction
Since the inception of machine learning, especially convolutional neural networks - they have been widely used in various fields of image processing – including in medical imaging. CNNs trained in a supervised manner to classify various diseases have shown their immense potential1; however, they typically come with the so-called “black-box” problem – as the complex reasoning done by such models are difficult to comprehend by humans. To address this problem, several post-hoc interpretability techniques can be applied2,3. On the other hand, CNNs can also be trained to segment relevant parts of the image, such as tumours. Typically, they are trained in a supervised manner and warrant a manually annotated dataset4. Such annotated datasets need to be prepared by clinicians, which is a time-consuming and daunting task. This work presents a deep learning-based classifier that inherently provides attributions on the input – to be able to understand which parts of the image had the highest impact on the model’s decision. Moreover, the potential of extending such attributions to perform segmentation without explicitly training the model with a manually segmented dataset to perform segmentation has also been shown.Methods
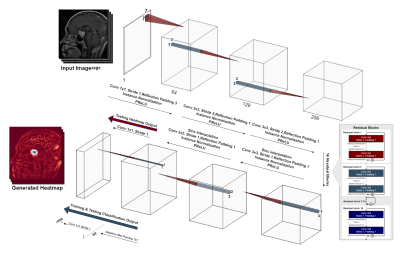
A typical deep learning-based model classification model, like VGG5, ResNet6 or InceptionNet7, condenses the input image using strided-convolutions or pooling layers repeatedly. Finally, the most condensed form, which might typically be multiple features, but just a single pixel per feature, is then forwarded to a fully-connected layer to obtain the final prediction. Contrastingly, a typical segmentation or even reconstruction model, such as U-Net8 or ReconResNet9, condenses the input in a similar fashion to obtain a latent representation, commonly known as the contraction operation. Then the latent representation is expanded multiple times using transposed-convolution or interpolation+convolution or even using unpooling layers to finally obtain the original input size back – commonly known as the expansion operation. GP-UNet10 combined these two techniques into one and used it to learn segmentation with regression labels. The current research extends that for brain tumour classification and segmentation using a different segmentation model, ReconResNet9: GP-ReconResNet.In this method (Fig. 1), initially, the input is given to a ReconResNet9 model (Fig. 2). This model provides n output images of the same size as the input, where n is the number of desired segmentation classes. On the contrary, the proposed method first sends the output to a global adaptive pooling operation to convert the full-size output into one pixel for each class and then sends it to the final fully-connected convolution layer, which also provides a single-pixel (neuron) for each class. Consequently, the network can be trained using classification image-level labels instead of segmentation labels. During inference, this adaptive max-pooling operation can be disabled, so the full-sized output of the transposed-convolution block can directly go to the fully-connected convolution layer to generate one output of the size of the input per class: known as the class activation maps or heatmaps, and by looking at them, the model’s focus areas can be understood without applying any post-hoc interpretability method. The adaptive max-pooling operation can be turned on again to obtain the classification result from the same model during inference. To generate the segmentation from this model, the generated heatmap is first suppressed (negative values set to 0) then thresholded with Otsu’s method with an offset of 0.1.
The proposed method has been evaluated on a brain tumour dataset containing 3064 T1-weighted contrast-enhanced MRIs, a mix of axial-coronal-sagittal orientations, from 233 patients with three kinds of brain tumours: meningioma (708 slices), glioma (1426 slices), and pituitary tumour (930 slices). The data was split into training, validation and testing sets, with a ratio of 60:15:25. Mixed-precision training was performed using weighted-cross-entropy loss and was optimised using the Adam optimiser for 150 epochs (until convergence) with a learning rate of 0.001
Results
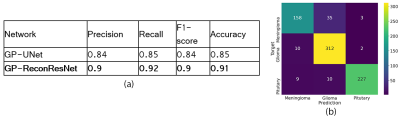
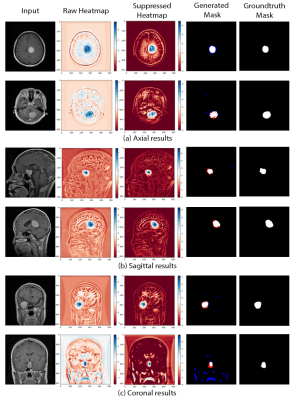
The proposed GP-ReconResNet achieved an F1-score of 0.9 on the test set. Fig. 3 shows the scores obtained on the evaluation metrics and were compared against an adapted version of the GP-UNet10 for this task. Fig. 4 shows the explainability results and the corresponding segmentations. To focus on the tumour, suppressed versions of the heatmaps were created - focusing on the positive attributions only, where blue signifies the main focus areas of the network. Finally, the segmentation masks and their comparisons with the ground-truth masks are shown.Discussion
The heatmaps make the GP-ReconResNet explainable, as they show the network’s focus area while predicting the tumour without applying any post-hoc techniques. Moreover, this research also shows the potential of performing tumour segmentation by post-processing the heatmaps - which does not require any manually annotated training data. Even though the segmentations are not perfect, it shows the potential of being part of a decision support system.Conclusion
This research proposed the GP-ReconResNet, which uses ReconResNet as the backbone model, combined with global pooling to classify the input image. The network’s focus area (heatmap) can be obtained by removing the pooling, making this network explainable. By thresholding the heatmaps, segmentation results can be obtained without training separately for segmentation. The applicability of this network and its superiority over using UNet as the backbone model have been shown for the task of tumour segmentation.Acknowledgements
This work was in part conducted within the context of the International Graduate School MEMoRIAL at OvGU (project no. ZS/2016/08/80646) and supported by the Wu Tsai Neurosciences Institute Seed Grant program.References
[1] Noor, Manan Binth Taj, et al. "Detecting neurodegenerative disease from mri: A brief review on a deep learning perspective." International Conference on Brain Informatics. Springer, Cham, 2019.
[2] Chatterjee, Soumick, et al. "Exploration of interpretability techniques for deep covid-19 classification using chest x-ray images." arXiv preprint arXiv:2006.02570 (2020).
[3] Chatterjee, Soumick, et al. "TorchEsegeta: Framework for Interpretability and Explainability of Image-based Deep Learning Models." arXiv preprint arXiv:2110.08429 (2021).
[4] Chatterjee, Soumick, et al. "Classification of Brain Tumours in MR Images using Deep Spatiospatial Models." arXiv preprint arXiv:2105.14071 (2021).
[5] Simonyan, Karen, and Andrew Zisserman. "Very deep convolutional networks for large-scale image recognition." arXiv preprint arXiv:1409.1556 (2014).
[6] He, Kaiming, et al. "Deep residual learning for image recognition." Proceedings of the IEEE conference on computer vision and pattern recognition. 2016.
[7] Szegedy, Christian, et al. "Rethinking the inception architecture for computer vision." Proceedings of the IEEE conference on computer vision and pattern recognition. 2016.
[8] Ronneberger, Olaf, et al. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
[9] Chatterjee, Soumick, et al. "ReconResNet: Regularised Residual Learning for MR Image Reconstruction of Undersampled Cartesian and Radial Data." arXiv preprint arXiv:2103.09203 (2021).
[10] Dubost, Florian, et al. "Gp-unet: Lesion detection from weak labels with a 3d regression network." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2017.
Figures