0168
An unsupervised deep learning method for affine registration of multi-contrast brain MR images1R&D, Subtle Medical Inc, Menlo Park, CA, United States
Synopsis
Image registration is a crucial preprocessing step for many downstream analysis tasks. Existing iterative methods for affine registration are accurate but time consuming. We propose a deep learning (DL) based unsupervised affine registration algorithm that executes orders of magnitude faster when compared to conventional registration toolkits. The proposed algorithm aligns 3D volumes from the same modality (e.g. T1 vs T1-CE) as well as different modalities (e.g. T1 vs T2). We train the model and perform quantitative evaluation using a pre-registered brain MRI public dataset.
Introduction
Inter and intra-patient image registration is a key step in many analysis tasks. Existing standard toolkits [1-4] are accurate but slow as they implement iterative optimization-based approaches which is a bottleneck to many existing medical image analysis pipelines. In the conventional iterative methods, the parameters are calculated from scratch for every image pair without utilizing prior information about the images. Fast affine registration is necessary as a prerequisite for deformable registration or by itself for non-deforming organs such as the brain. We propose a DL-based affine registration algorithm by building on an existing framework [5] for deformable image registration. We also perform quantitative evaluation utilizing a publicly available pre-registered dataset.Methods
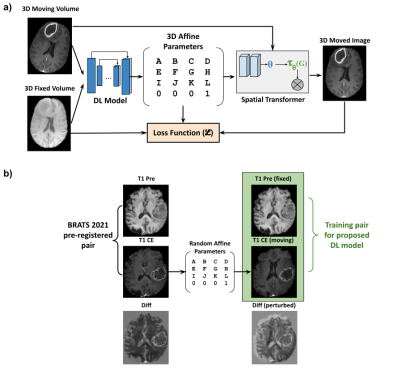
Unsupervised registrationFigure 1a depicts the premise of the proposed work which was based on an existing unsupervised deformable registration framework - Voxelmorph [5]. The fixed image (template image) and the moving image (image to be registered) were passed through a DL network which extracted the image features and predicted the affine parameters. These affine parameters were passed through a spatial transformer layer [6] which essentially is a differentiable module that applies the affine transformation on the input image. The output of the spatial transformer layer (moved image) was subject to a loss function with respect to the fixed image.
Dataset
We used the publicly available BRATS 2021 dataset [7-9] which consisted of 1251 cases with T1, T1-CE, T2 and FLAIR images. The cases also had the ground truth tumor segmentations. The brain images were skull-stripped and pre-registered to an anatomical template. We used 1126 cases for training and 125 for validation and quantitative evaluation of registration performance.
Training pair generation
Figure 1b shows the process of training pair generation. The T1-weighted pre-contrast volumes were used as the fixed template image while the corresponding T1-CE volumes were subject to random affine transformations which were within a predefined range. The input volume pair for the DL model training was the T1 pre-contrast volume (fixed) and the randomly affine transformed T1-CE volume (moving).
Network Architecture
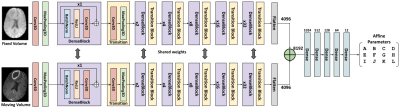
Figure 2 shows the proposed multi-path residual dense network. The fixed and the moving volumes have separate encoder pathways but with shared weights. This proposed network architecture was a modified version of [10]. The input volume was first passed through a 3D convolution layer followed by a max-pooling layer. Followed by this, the encoder consisted of six consecutive pairs of dense blocks [11] and transition blocks. The depth of the dense block was progressively increased by a factor of 2n. The feature maps in the respective pathways were then flattened and concatenated and passed through a series of fully connected layers to finally output the 12 affine parameters. This network was trained end-to-end in conjunction with the Voxelmorph framework shown in Fig 1b.
Experiments
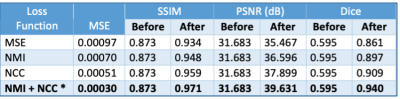
Three models were trained with MSE, normalized cross-correlation (NCC) [12] and normalized mutual information (NMI) [13] as loss functions. Adam optimizer was used with a learning rate of 1e-4 combined with a scheduler where the learning rate was halved every 50 epochs. A fourth model was also trained with a combination of NCC and NMI losses. The same scheme was extended to train a model to register T2 and FLAIR images with T1 as the reference image.
Quantitative evaluation
The four models were quantitatively evaluated and compared using the following metrics:
(1) Mean Squared Error (MSE)
MSE was computed between the inverse of the simulated random affine transformation matrix and the mode predicted affine matrix.
(2) SSIM & PSNR
Structural similarity metric (SSIM) and peak signal-to-noise ratio (PSNR) was computed between the registered image predicted by the corresponding models and the pre-registered image available in the BRATS dataset.
(3) Dice score
Let $$$M$$$ be the binary tumor segmentation mask available in the BRATS dataset. Let $$$\theta_{sim}$$$ be the simulated random affine parameters and let $$$\theta_{pred}$$$ be the affine transformation predicted by the DL model. We first transformed $$$M$$$ with $$$\theta_{sim}$$$ - the tumor mask on the perturbed volume; we then transformed the resultant perturbed mask with $$$\theta_{pred}$$$ and call it $$$\hat{M}$$$ which essentially "registers" the perturbed mask; we have
$$\hat{M} = M . \theta_{sim} . \theta_{pred}$$
We evaluate the registration performance by computing $$$Dice(M,\hat{M})$$$.
Results
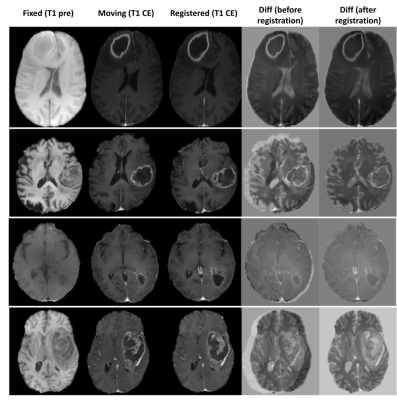
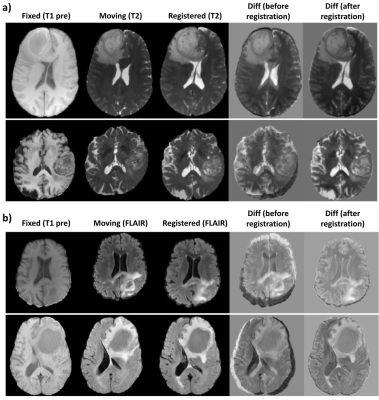
Figure 3 shows the quantitative results of the different models.The combination of NCC and NMI losses yielded the best results. When compared to SimpleElastix [14] (with default settings) which took 24 seconds for registering a pair of 128x256x256 volumes, the proposed model takes only ~0.8 seconds for the same. Figure 4 shows sample qualitative results of T1 vs T1-CE registration. Figures 5a&b show qualitative results for cross-modality registration of T1 vs T2 and T1 vs FLAIR respectively.Discussions
Based on the quantitative assessment and qualitative results, we have shown that DL models can be used for fast and accurate affine registration of multi-modal brain MRI images. The same scheme can be extended for registering cross modal (PET/CT, PET/MR, MR/CT) image volumes. An end-to-end network can also be developed that performs affine and non-rigid registration sequentially.Conclusion
We have proposed a fast and accurate unsupervised DL approach for affine registration of brain MRI images.Acknowledgements
We would like to acknowledge the grant support of NIH R44EB027560.References
B. C. Lowekamp, D. Chen, I. Luis, and B. Daniel. The design of simpleitk. Frontiers in Neuroinformatics, 7(7):45, 2013.
Z. Yaniv, B. C. Lowekamp, H. J. Johnson, and R. Beare. Simpleitk image-analysis notebooks: a collaborative environment for education and reproducible research. Journal of Digital Imaging, 31(3):1–14, 2017.
B. B. Avants, N. J. Tustison, G. Song, P. A. Cook, A. Klein, and J. C. Gee. A reproducible evaluation of ants similarity metric performance in brain image registration. Neuroimage, 54(3):2033–2044, 2011.
A. Fedorov, R. Beichel, J. Kalpathy-Cramer, J. Finet, J. C. Fillion-Robin, S. Pujol, C. Bauer, D. Jennings, F. Fennessy, and M. Sonka. 3D Slicer as an image computing platform for the quantitative imaging network. Magnetic Resonance Imaging, 30(9):1323–1341, 2012.
Guha Balakrishnan, Amy Zhao, Mert R. Sabuncu, John Guttag, Adrian V. Dalca. VoxelMorph: A Learning Framework for Deformable Medical Image Registration. IEEE TMI: Transactions on Medical Imaging. 2019. eprint arXiv:1809.05231
M. Jaderberg, K. Simonyan, A. Zisserman, and K. Kavukcuoglu. Spatial transformer networks. Advances in Neural Information Processing Systems, 28:2017–2025, 2015.
U.Baid, et al The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. arXiv:2107.02314, 2021.
B. H. Menze, A. Jakab, S. Bauer, J. Kalpathy-Cramer, K. Farahani, J. Kirby, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS), IEEE Transactions on Medical Imaging 34(10), 1993-2024 (2015) DOI: 10.1109/TMI.2014.2377694
S. Bakas, H. Akbari, A. Sotiras, M. Bilello, M. Rozycki, J.S. Kirby, et al., Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features, Nature Scientific Data, 4:170117 (2017) DOI: 10.1038/sdata.2017.117
Evelyn Chee and Zhenzhou Wu. AIRNet: Self-Supervised Affine Registration for 3D Medical Images using Neural Networks. arXiv 1810.02583; 2018.
G. Huang, Z. Liu, L. V. D. Maaten, and K. Q. Weinberger. Densely connected convolutional networks. In IEEE Conference on Computer Vision and Pattern Recognition, pages 2261–2269, 2017.
Kaso A. Computation of the normalized cross-correlation by fast Fourier transform. PLoS ONE 13(9): e0203434. https://doi.org/10.1371/journal.pone.0203434.
M Hoffmann, B Billot, JE Iglesias, B Fischl, AV Dalca. Learning image registration without images. arXiv preprint arXiv:2004.10282, 2020. https://arxiv.org/abs/2004.10282
Kasper Marstal, Floris Berendsen, Marius Staring and Stefan Klein, SimpleElastix: A user-friendly, multilingual library for medical image registration, International Workshop on Biomedical Image Registration (WBIR), 2016
Figures