0159
MRI tracking of genetically engineered extracellular vesicles as SARS-CoV-2 mimetics for studying viral-induced pathologies
Andrea Galisova1, Jiri Zahradnik1, Hyla Allouche-Arnon 1, Gideon Schreiber1, and Amnon Bar-Shir1
1Weizmann Institute of Science, Rehovot, Israel
1Weizmann Institute of Science, Rehovot, Israel
Synopsis
Extracellular vesicles (EVs) are cell derived nanoformulations that allow to use genetic tools to engineer their features. Here, we show an MR imaging platform, which is based on genetically engineered magnetically labeled EVs, for mapping the binding of SARS-CoV-2 mimetics to the ACE2 receptor in vivo. The proposed imaging platform can help to better understand the mechanism and development of SARS-CoV-2 infection and in the delivery of proposed therapeutics. The presented principles can be further extended to study other viruses and viral mutations by engineering a tailored peptide on the EVs surface and thus study viral-induced pathologies by MRI.
Introduction
Visualization of viral pathways in an infected subject and understanding viral-induced pathologies is of great interest both scientifically and therapeutically, particularly in the era of the COVID-19 pandemic. In this regard, extracellular vesicles (EVs) offer a unique platform for studying viral infection paths and mechanisms due similar size and biomembrane properties as viruses [1]. In our lab we developed genetically engineered EVs displaying the receptor-binding domain (RBD) of SARS-CoV-2 on their surface creating non-infectious corona-virus-like mimetics (EVsRBD) binding to the ACE2 receptor. Importantly EVs can be labeled magnetically (e.g. with superparamagnetic iron oxide nanoparticles, SPIONs) [2, 3] and visualized by real-time MRI using T2/T2*-weighted images. Here we show MRI visualization of in vivo targetability of the EVsRBD in a mouse model, which can be further extended to study the manifestation of the SARS-CoV-2 virus or other viruses in different organs in real time.Methods
HEK293 cells were genetically modified to express the RBD of SARS-CoV-2 on their surface (Fig. 1a). The secreted EVs – SARS-CoV-2 mimetics referred here as EVsRBD (or EVsnoRBD as controls) were isolated by differential centrifugation. Binding of the EVsRBD to the ACE2 receptor was examined after incubation of EVsRBD with the ACE2-expressing cells by fluorescence imaging and flow cytometry. For MRI, the parental cells were incubated with SPIONs (BioPal, 2.5-40 µg/mL) for 24 hours (Fig. 2a) and the secreted SPIONs-labeled EVs were characterized by western blot for protein expression, nanoparticle tracking for their size and by T2-weighted MRI (RARE sequence, TR=2800 ms, TE=42 ms, resolution 0.23×0.23×1 mm3). Two types of subcutaneous tumors were induced in nude mice (HEK293T as control or HEK293T overexpressing ACE2) to study in vivo targeting of EVsRBD to the ACE2 receptor. Genetically engineered EVs (3x1011), either presenting the RBD (EVsRBD) or control (EVsnoRBD) labeled with SPIONs for MRI or with DiR for fluorescence were intravenously injected followed by in vivo MRI (15.2T MR scanner) and ex vivo fluorescence imaging (IVIS Lumina XR). T2-/T2*-weighted MR images of mice were acquired before and after intravenous administration of the SPIONs-labeled EVs (FLASH sequence, TR=300 ms, TE=2 ms, resolution 0.23×0.23×0.7 mm3). To confirm the presence of iron, tumor tissues were processed by histology (Prussian Blue staining).Results
Following incubation of the HEK293 cells with SPIONs, the secreted genetically engineered EVs showed expression of RBD (for EVsRBD) (Fig. 2c) and typical EVs size (~100 nm) (Fig. 2d). Binding to the ACE2 receptors was confirmed by higher uptake of EVsRBD in ACE2-expressing cells compared to control EVsnoRBD (Fig.1b, c). MRI of isolated EVs showed concentration-dependent accumulation of SPIONs as depicted from the lower MRI signal on T2-weighted images (Fig. 2e, f) confirming successful labeling of the engineered EVs, which was furthered confirmed by TEM images (Fig. 2b). In vivo targetability of EVsRBD was demonstrated by both MRI and fluorescence imaging of mice after EVs injection. The higher accumulation of the EVsRBD in ACE2 tumor as compared to control was observed by the lower MRI signal (T2*-weighted images) of tumor tissue (Fig. 3a,b). Ex vivo fluorescence imaging (Fig. 3c) and histology examination of tumors confirmed the higher accumulation of EVsRBD in the ACE2 tumors compared to EVsnoRBD (Fig. 3d).Conclusion
Developing genetically engineered, magnetically labeled EVs, we showed here the ability to use MRI for mapping the binding of coronavirus mimetics to the ACE2 receptor in vivo. The principles described in this study could be further extended to better understand the mechanism and development of SARS-CoV-2 infection and the proposed platform can be implemented to study other viruses by engineering a tailored peptide on the surface of EVs using genetically engineering principles and thus to explore the role of receptors in a wide spectrum of viral-induced pathologies.Acknowledgements
This study was supported by the Ben B. and Joyce E. Eisenberg Foundation.References
[1] Hoen, EN, et al. PNAS 2016, 113(33) [2] Han Z, et al. Journal of Extracellular vesicles 2021, 10(3): e12054 [3] Busato, A, et al. International Journal of Nanomedicine 2016, 11:2481-2490Figures
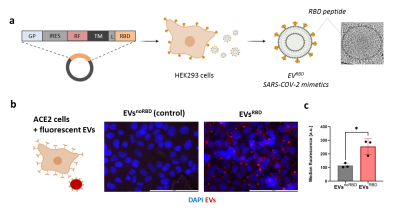
Fig. 1. Characterization of SARS-CoV-2 mimetics. (a) Engineering SARS-CoV-2 mimetics by genetic modification of parental cells. The released EVs present on their surface the receptor binding domain (RBD) of SARS-CoV-2 virus. The shape and size of EVs was confirmed by TEM. (b) Targeting of EVsRBD to the ACE2 receptor was confirmed by incubation of EVs with ACE2-expressing cells. (c) FACS analysis showed enhanced accumulation of EVsRBD compared to EVsnoRBD in ACE2 expressing cells.
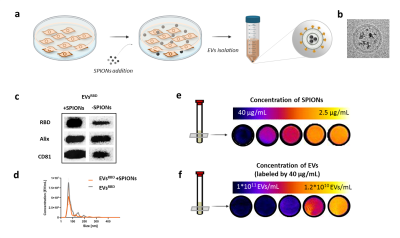
Fig. 2. Characterization of magneticaly-labeled EVs. (a) Schematic illustration of EVs labeling with SPIONs by incubating the parental cells with the nanoparticles. (b) TEM image of isolated magnetically-labeled EV confirming the presence of SPIONs inside EVsRBD. (c) Protein expression of peptides and (d) size of SPIONs labeled and unlabeled EVs is similar. T2-weighted MRI signal of magnetically-labeled EVs is (e) proportional to the concentration of SPIONs used for labeling and (f) to concentration of EVs in the sample.
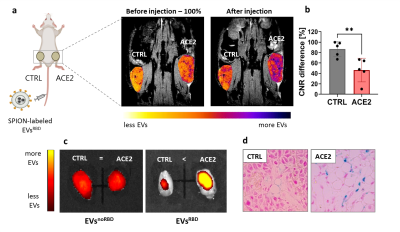
Fig. 3. In vivo MRI and optical imaging tracking of EVsRBD. (a) In vivo MRI imaging of tumor-bearing mice showed preferential accumulation of EVsRBD in the ACE2-expressing tumors compared to the control tumors. (b) T2*-weighted images of mice show lower MR signal in the control tumors after injection of EVsRBD. (c) Ex vivo fluorescence imaging of tumors confirmed higher accumulation of EVsRBD in ACE2 tumors compared to the control vesicles (EVsnoRBD). (d) Accumulation of magnetically labeled EVs in the ACE2 tumors was by histology using Prussian Blue staining for iron content.
DOI: https://doi.org/10.58530/2022/0159