0149
Assessment and improvement of the quality of fat saturation in breast MRI using deep-learning with synthetic data1Division of biomedical engineering, Hankuk university of foreign studies, gyunggi-do,yongin-si, Korea, Republic of, 2Department of Radiology, Seoul St. Mary's Hospital, Seoul, Korea, Republic of
Synopsis
We propose a fully automatic method to assess and improve the quality of the fat saturation in breast MR images. For this purpose, three deep neural networks were trained using both actual and synthetic breast MR data. Firstly, the poorly fat saturated cases were classified using a binary classification network. Then, the poorly fat saturated regions were localized using a segmentation network. Lastly, for the poor cases, the remaining fat signals were retrospectively suppressed using a correction network. The results showed that our networks successfully identified the poor cases and suppressed the remaining fat signals.
Introduction
Accurate fat saturation(FS) in the breast MRI is important for diagnosis of breast cancer.1 However, FS using spectral selective RF pulses often fails or is insufficient due to field inhomogeneities.2 In breast MRI, this heterogeneity occurs more often in the relatively large breasts and the posterior aspect of the breast and in the axilla.3 If the fat signal is not sufficiently saturated, it can negatively affect evaluation of a breast cancer. Therefore, the quality of FS is important for accurate diagnosis of breast cancer. In this study, we propose a deep learning-based automatic method to assess and improve the quality of FS in breast MRI.Methods
[Dataset]In this study, T1-weighted MR images of 239 patients with breast cancer were retrospectively collected. By observing the maximum intensity projection images, a similar number of cases with relatively good FS and cases with poor FS were collected. For training the model, the quality of the FS was semi-automatically classified into two labels (0: good, 1: poor). By reviewing cross-sectional images, 103 cases out of 239 were labeled as poor FS. The MR images were acquired at 3T clinical scanner (Siemens Verio) using fat suppressed 3D VIBE sequences.
[Network]
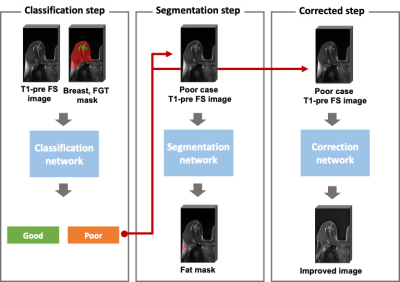
Our proposed method consists of three deep learning-based models (Figure 1). To assess the quality of the FS, two independent deep learning models were trained. The first model was to distinguish poor cases, and an image-level classification model using the two labels (good and poor) was trained. The second model was to segment the insufficiently saturated fat signals, and a segmentation model was trained. The VGG-16 was used for a classification task and the U-Net was used for a segmentation task, respectively.4,5
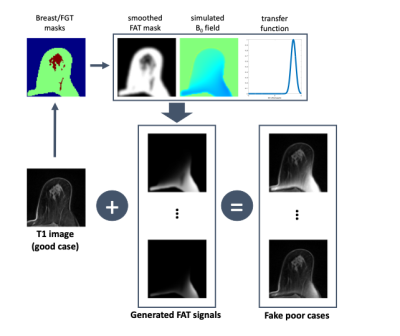
To improve the quality of the FS for the poor cases, a deep learning model was trained as a third model. Because we didn’t have paired data for good and poor, we generated the realistic fake poor cases from the good cases through magnetic field simulations as shown in Figure 2. Firstly, the MR images were segmented into the fat and fibroglandular tissue (FGT) regions using the previously developed method.6 Second, a magnetic field was calculated by assigning a susceptibility difference of 9 ppm between the whole breast and the background.7 Lastly, using the Gaussian transfer function for the B0 offset, virtual remaining fat signals were generated from the fat mask and added to the original T1 image. Using the generated poor cases as input data and the corresponding good cases as output data, the 3D U-Net was trained in a supervised manner. 5
[Evaluation]
Five fold cross validation was used to train and evaluate the neural networks. For the binary classification results (good or poor), the accuracy, F1 score, and receiver operating curve (ROC) were calculated. For the segmentation results, the true positive rate (TPR) was calculated. The correction model trained with the synthesized paired data was visually evaluated using both the synthetic poor cases and the actual poor cases.
Results and Discussion
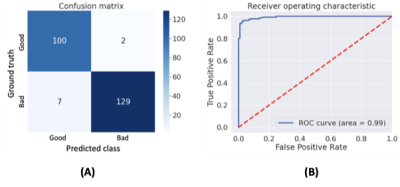
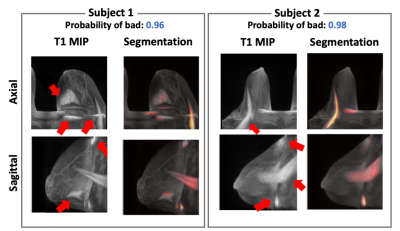
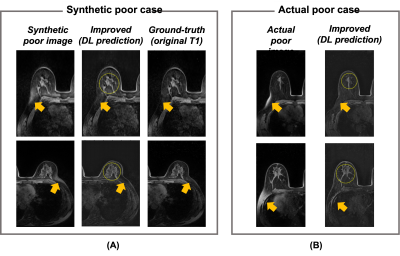
Figure 3 summarizes the classification results for the quality of FS. The calculated accuracy and F1 score were 96.2% and 0.96, respectively. The calculated TPR from the segmentation results was 74.4%. Figure 4 shows the representative results for the segmentation result of the unsaturated fat signals. Figure 5 shows the test results using the quality improvement network for the synthetic poor cases and actual poor cases. The proposed method effectively suppressed the remaining fat signals (arrows in Fig. 5) while maintaining the FGT signals (circles in Fig. 5) in most cases (both the synthetic poor cases and actual poor cases).Conclusion
In this study, we proposed the deep learning-based method for assessing and improving the quality of FS in breast MRI. The method successfully identified the poor FS cases and localized the remaining fat signals. In addition, the remaining fat signals were retrospectively suppressed by the model trained by the synthetic images. To show potential value of the proposed method, further validation studies considering the clinical situations are needed.Acknowledgements
This work was supported by the Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, Republic of Korea, the Ministry of Food and Drug Safety) (Project Number: 202011C20)References
1. Lin C, Rogers CD, Majidi S. Fat suppression techniques in breast magnetic resonance imaging: A critical comparison and state of the art. . 2015.
2. Delfaut EM, Beltran J, Johnson G, Rousseau J, Marchandise X, Cotten A. Fat suppression in MR imaging: Techniques and pitfalls. Radiographics. 1999;19(2):373-382.
3. Dogan BE, Ma J, Hwang K, Liu P, Yang WT. T1‐weighted 3D dynamic contrast‐enhanced MRI of the breast using a dual‐echo dixon technique at 3 T. Journal of Magnetic Resonance Imaging. 2011;34(4):842-851.
4. Kaiming He, Xiangyu Zhang, Shaoqing Ren, Jian Sun. Deep residual learning for image recognition. CVPR. Jun 2016:770-778. https://ieeexplore.ieee.org/document/7780459. doi: 10.1109/CVPR.2016.90.
5. Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-net: Learning dense volumetric segmentation from sparse annotation. . 2016:424-432.
6. Nam Y, Park GE, Kang J, Kim SH. Fully automatic assessment of background parenchymal enhancement on breast MRI using Machine‐Learning models. Journal of Magnetic Resonance Imaging. 2021;53(3):818-826.
7. Jordan CD, Daniel BL, Koch KM, Yu H, Conolly S, Hargreaves BA. Subject‐specific models of susceptibility‐induced B0 field variations in breast MRI. Journal of Magnetic Resonance Imaging. 2013;37(1):227-232.
Figures