0047
The Open Science Initiative for Perfusion Imaging (OSIPI): Early Results from the DCE-MRI Challenge1School of Physics and Astronomy, University of Leeds, Leeds, United Kingdom, 2The University of Alabama at Birmingham, Birmingham, AL, United States, 3Department of Radiology & Nuclear Medicine, Erasmus MC University Medical Center, Rotterdam, Netherlands, 4Mayo Clinic, Rochester, MN, United States, 5Department of Radiology, Neuroradiology Division, Mayo Clinic, Scottsdale, AZ, United States, 6Oden Institute for Computational Engineering and Sciences, The University of Texas at Austin, Austin, TX, United States, 7University of Texas at Austin, Austin, TX, United States, 8Neurological Imaging, Barrow Neurological Innovation Center, Phoenix, AZ, United States, 9Department of Surgery and Cancer, Imperial College London, London, United Kingdom, 10Department of Biomedicine and Prevention, University of Rome “Tor Vergata”, Roma, Italy, 11Department of Surgery & Cancer, Imperial College London, London, United Kingdom, 12Department of Medical Physics, Royal Surrey NHS Foundation Trust, Guildford, United Kingdom, 13Department of Biomedicine and Prevention, University of Rome "Tor Vergata", Rome, Italy, 14Athinoula A. Martinos Center for Biomedical Imaging, Harvard Medical School, Boston, MA, United States, 15University Medicine Göttingen, Göttingen, Germany, 16Indian Institute of Technology Delhi, New Delhi, India, 17Singapore Bioimaging Consortium (SBIC), Singapore, Singapore, 18GPU.IO, Pune, India, 19Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States, 20Quantitative MR Imaging and Spectroscopy Group, Research Center for Molecular and Cellular Imaging, Tehran University of Medical Sciences, Tehran, Iran (Islamic Republic of), 21Centre for Computational Imaging & Simulation Technologies in Biomedicine, School of Computing / School of Medicine, University of Leeds, Leeds, United Kingdom, 22Neuroradiology Division, Department of Radiology, Mayo Clinic, Phoenix, AZ, United States, 23Genentech, Inc., South San Francisco, CA, United States, 24Department of Infection, Immunity and Cardiovascular Disease, University of Sheffield, Sheffield, United Kingdom, 25Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States
Synopsis
While there is growing evidence that DCE-MRI may provide insights about the response of patients to therapies, there is a lack of standardized software quantification tools, resulting in variability in reported Ktrans values across different studies and limiting its utility in clinical applications. We have designed and launched the Open Science Initiative for Perfusion Imaging (OSIPI)-DCE challenge to provide recommended and benchmarked analysis tools for Ktrans estimation in the brain, by evaluating and comparing DCE software tools in terms of accuracy, repeatability, and reproducibility. Here, we report on the preliminary results of this challenge.
Purpose
There is growing evidence suggesting that volume transfer constant (Ktrans) quantified from dynamic contrast-enhanced (DCE-) MRI may provide helpful insights about response of brain tumors to treatments 1. It may therefore serve as a surrogate biomarker for characterizing tumor recurrence 1,2. However, the utility of this method is currently limited in clinical applications, due to a lack of standardized analysis tools, resulting in reported Ktrans variability between studies 3. Mounting concerns regarding the reproducibility of MRI scientific research have led to several attempts in standardizing analysis tools through community challenges, to provide validation and benchmarking quantification within a controlled setting 4,5. In the OSIPI-DCE challenge 6,7, we aim to validate and compare the analysis pipelines to estimate Ktrans in terms of their accuracy, repeatability, and reproducibility in a controlled setting. The results of this ongoing challenge will provide guidelines about the factors to consider for quantification of Ktrans in brain tumors and an opportunity for benchmarking future software development by offering access to the challenge data and scoring metrics. In this abstract, we report the early results for OSIPI-DCE challenge, providing a preliminary analysis and comparison of the received submissions in terms of accuracy and repeatability.Methods
Challenge Setup. The OSIPI-DCE challenge was launched in May 2021 and will accept submissions through December 2021. Many researchers in the field of contrast-based perfusion MRI were invited to visit the ISMRM challenge website 8 and participate in the challenge. The interested participants submitted their results through the website 8, including their source codes/software, Ktrans maps in NIfTI format, and an accompanying standard operating procedure (SOP) containing sufficient details for a neutral team to reproduce the results without additional information.Data Description. Two sets of data were provided 6,7: (1) clinical: a set of test-retest DCE-MRI and T1-mapping scans from eight patients with brain tumors; and (2) synthetic: two synthetic patient DCE-MRI datasets with test-retest scans, generated from clinical subjects to produce ground truth data that mimic actual data and can be analyzed with the same processing pipeline as the clinical data.
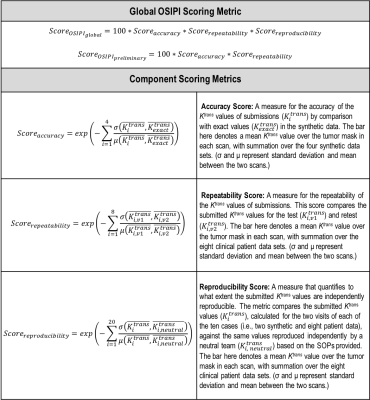
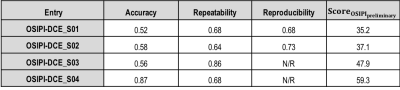
Evaluation Process. Segmentation of brain tumors was performed on DCE-MRI scans of clinical and synthetic data in two test-retest visits using ITK-SNAP software 9. The segmentation mask was overlaid on the ground truth and submitted Ktrans maps in NIfTI format 10. Mean Ktrans were calculated within the tumor masks, with normalised standard deviation in mean tumor values between visits for clinical patients or, between submitted and ground truth Ktrans for synthetic cases. Scoring metrics for accuracy, repeatability, and reproducibility (as defined in Table 1) were used to define an OSIPI score for submissions.
Results
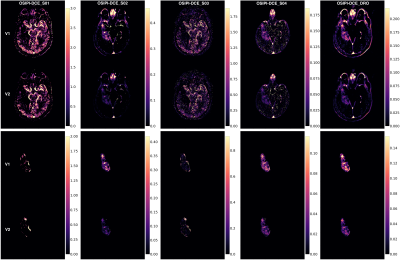
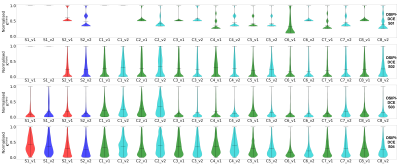
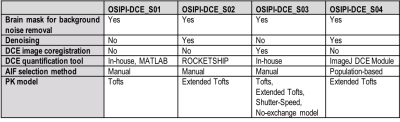
Challenge Entries. For this early report on the challenge, we evaluated four received submissions, identified with pseudo-IDs, OSIPI-DCE_S01-4. A summary of the methods including preprocessing methods (brain masking, denoising, co-registration), PK model, AIF selection method, and the DCE quantification tool used for each submission is provided in Table 2.Scoring. The scores for the four received submissions are shown in Table 3, with entry OSIPI-DCE_S04 scoring the highest in all areas. The Ktrans maps from the synthetic data for all submissions are compared to the ground truth, shown in Figure 1 for a singular slice with synthetic patient 2. Here, the bottom panel shows the submissions and ground truth after application of the analysis tumor masks. The tumor regions between submissions show similar intensity layout but have highly variable scaling in magnitude. The normalised Ktrans distributions for the entries are illustrated in Figure 2 for comparison. This shows visually detectable distribution variation between some test-retest patients for comparable mean values. The reproducibility test requires independent replication of the SOPs by a neutral team, and at this time, we provide preliminary results for the first two challenge entries as an example. As the metric is not available for all entries, it is not considered in calculation of preliminary OSIPI score (ScoreOSIPI_preliminary) in Table 3. Upon closure of the challenge in the near future, we will provide a full report of reproducibility by applying this test to all challenge submissions in calculation of “ScoreOSIPI_global”.
Discussion and Conclusion
The preliminary results of validation and benchmarking the early received submissions suggest large variability of Ktrans estimations across different submissions. Overall, the repeatability was higher than the accuracy, indicating that Ktrans could be more effective for therapy monitoring rather than diagnosis. Some entries used non-standard DICOM-to-NIfTI conversion and required realignment of these submissions to ensure overlap with the tumor masks, which may have produced rounding errors, impacting the calculated scores. We predict that independent reproducibility of the submissions with manual AIF selection might be challenging. The final challenge analysis will be developed from the methodology presented here, this preliminary analysis has been invaluable for testing the scoring metrics robustly. The scoring metric will be refined to (1) calculate metrics for smaller regions of interest within tumor masks; and (2) normalize the OSIPI score for each entry with respect to a standard model to improve interpretability. Looking forward, we hope these results encourage more submissions; our metrics will determine whether any can outperform the current highest-ranking entry (OSIPI-DCE_S04) in terms of accuracy and repeatability.Acknowledgements
No acknowledgement found.References
1. Alic, L., van Vliet, M., Van Dijke, C. F., et al. Heterogeneity in DCE-MRI parametric maps: a biomarker for treatment response?. Phys Med Biol, 56(6), 1601 (2011).
2. Elshafeey, N., Kotrotsou, A., Hassan, A. et al. Multicenter study demonstrates radiomic features derived from magnetic resonance perfusion images identify pseudoprogression in glioblastoma. Nat Commun 10, 3170 (2019). https://doi.org/10.1038/s41467-019-11007-0
3. Okuchi, S., Rojas-Garcia, A., et al. Diagnostic accuracy of dynamic contrast-enhanced perfusion MRI in stratifying gliomas: A systematic review and meta-analysis. Cancer Med. 8(12):5564-5573 (2019).
4. Stikov, N., Trzasko, J. D. & Bernstein, M. A. Reproducibility and the future of MRI research. Magn. Reson. Med. 82, 1981–1983 (2019).
5. Maier, O., Baete, S. H., Fyrdahl, A., et al. CG‐SENSE revisited: Results from the first ISMRM reproducibility challenge. Magn Reson Med, 85(4), 1821-1839 (2021).
6. Kazerooni, A. F., Bell, L. C., et al. The Open Source Initiative for Perfusion Imaging (OSIPI): DCE-MRI Challenge, Intl. Soc. Magn. Reason. Med. (2021)
7. Kazerooni, A. F., Shalom, E., et al. (2021). OSIPI_TF6.2. https://doi.org/10.17605/OSF.IO/U7A6F
8. https://challenge.ismrm.org/forums/topic/osipi-dce-challenge/
9. Yushkevich, P. A., Piven, J., et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage Jul 1;31(3):1116-28 (2006).
10. Brett, M., Markiewicz, C.J., et al. nipy/nibabel: 3.2.1 2020. doi:10.5281/zenodo.4295521.
Figures