0016
A disentangled representation trained for joint reconstruction and segmentation of radially undersampled cardiac MRI
Tobias Wech1, Julius Frederik Heidenreich1, Thorsten Alexander Bley1, and Bettina Baeßler1
1Department of Diagnostic and Interventional Radiology, University Hospital Würzburg, Würzburg, Germany
1Department of Diagnostic and Interventional Radiology, University Hospital Würzburg, Würzburg, Germany
Synopsis
The network we propose in this work (xSDNet) jointly reconstructs and segments cardiac functional MR images which were sampled below the Nyquist rate. The model is based on disentangled representation learning and factorizes images into spatial factors and a modality vector. The achieved image quality and the fidelity of the delivered segmentation masks promise a considerable acceleration of both acquisition and data processing.
Purpose
We propose a model based on disentangled representation learning [1], which was trained to simultaneously reconstruct and segment radially undersampled cardiac functional MRI data with high fidelity and short runtimes.Methods
Model architectureFigure 1 provides a schematic overview of the proposed architecture, which was derived from the spatial decomposition network (SDNet) as presented in [1]. SDNet allows a factorization of medical image data into spatial factors (i.e. information associated with anatomy) and a modality vector, which encodes contrast information specific to the technique used for imaging (e.g. CT, MR, or a special contrast of one modality). This very general approach not only is attractive due to semantically meaningful latent factors, it is furthermore particularly suited to be trained for multi-task objectives. So far, applications cover segmentation of (fully sampled) cardiac MR images or the synthetization of plausible MR data in a generative fashion.
Our model, dubbed xSDNet, represents an extended version of SDNet (see Fig. 1), allowing both reconstruction and segmentation of cardiac functional MR images, which were sampled below the Nyquist rate. In essence, the output of the FiLM layers (Feature-wise linear modulation, [2]) of SDNet was additionally stacked with the undersampled data to build the input of the final UNet layers, ultimately yielding the desired images of high quality.
Data
Openly available radial raw data (multicoil, complex valued) of mid-ventricular cardiac functional exams [3,4] were used to train, validate and test the proposed model. This set was acquired in breath-hold using a fully sampled radial bSSFP sequence at 3T (TR = 3.1 ms, TE = 1.4 ms, in-plane resolution = 1.8 mm × 1.8 mm, slice thickness = 8 mm, FA = 48°, number of channels = 16 ± 1). Retrospective ECG-triggering was used to determine 25 cardiac phases in a segmented fashion, each consisting of 196 linearly ordered projections. 83 cine series of these data – each acquired in a different subject - were divided into 61 series for training (1525 images), 5 for validation (125 images) and 17 for testing (425 images).
Segmentation labels for left ventricle (LV), myocardium (MYO) and right ventricle (RV) were automatically determined for each frame of each cine series using the 2D model provided by Bai et al. [5].
Subsequently, sub-Nyquist sets of the raw data were created to simulate accelerated imaging by reconstructing only $$$ p \in \{98, 49, 33, 25\}$$$ projections per cine frame.
Training and Evaluation
xSDNet was trained using undersampled cine frames as input and both the determined segmentation masks and the according fully sampled reference frames as outputs. While remaining losses were used in accordance with the original SDNet, a perceptional loss was used for the reconstruction path in our approach.
For comparison, a classical 2D UNet was additionally trained for reconstructing the undersampled data, very similar to the benchmark presented in [6] (l2-loss). Both the latter and xSDNet were then applied to each image of the test series, separately for all undersampling rates as listed above.
One frame of each reconstructed series was subjected to an assessment of the image quality. Two expert radiologists in cardiac imaging, blinded for both reconstruction method and sampling rate, evaluated the following categories on a 5-point Likert scale: Spatial resolution (1-poor to 5-high), artifact level (1-severe to 5-no), contrast between myocardium and blood (1-poor to 5-high), signal-to-noise-ratio (1-low to 5-high), overall image impression (1-poor to 5-excellent).
The quality of the segmentation masks as second output of xSDNet was assessed by calculating mean Sørensen–Dice coefficients with respect to the reference masks as delivered by Bai’s model on fully sampled cines.
Results
Image reconstruction and segmentation of a single 2D cine frame took on average 19 ms when using xSDNet on an Nvidia Titan XP GPU. In Fig. 2, exemplary images are depicted for one subject and different undersampling factors. UNet reconstructions of the same data as well as fully sampled reference images are presented for comparison. Fig. 3 provides a dynamic view for a different patient, and Fig. 4 illustrates the same subject, with segmentation masks additionally superimposed. According to the results of the reader study (Tab. 1, left), xSDNet outperforms the benchmark UNet for all acceleration factors. For $$$p$$$ = 98, both models are only slightly inferior to the fully sampled reference. For $$$p$$$ < 98, the ratings of UNet drop rapidly, while xSDNet still performs robust. In particular, residual undersampling artifacts start to deteriorate image quality for xSDNet at $$$p$$$ = 25. Dice scores were high (see Tab. 1, right) and decrease only slightly when reducing the number of projections per frame.Discussion
The presented xSDNet promises a considerable acceleration of cardiac functional MRI, both in terms of acquisition and data processing. Applying the proposed model based on disentangled representation learning, joint reconstruction and segmentation of the undersampled images took less than half a second for a series consisting of 25 frames. Image quality and fidelity of the segmentation masks was high, even for only 33 projections per frame. Further improvement of the robustness and a generalization to basal and apical slices requires an adequate extension of the existing training dataset.Acknowledgements
The project underlying this report was funded by the German Federal Ministry of Education and Research (BMBF grant no. 05M20WKA). We thank Wenjia Bai and coauthors [5] and Hossam El-Rewaidy and coauthors [4] for providing their data and/or models for scientific studies. We further thank Spyridon Thermos for providing a pytorch implementation of SDNet (https://github.com/spthermo/SDNet), which served as a baseline for our method.References
[1] Chartsias et al. Med Image Anal 2019;58:101535. [2] Perez et al. AAAI 2018 3942-3951 [3] El-Rewaidy et al. Harvard Dataverse, https://doi.org/10.7910/DVN/CI3WB6 [4] El-Rewaidy et al. Magn Reson Med. 2021;85:1195-1208. [5] Bai et al. J Cardiovasc Magn Reson. 2018;20:65. [6] Zbontar et al. arXiv, https://arxiv.org/abs/1811.08839Figures
Figure 1: Illustration of the network architecture of the proposed xSDNet.
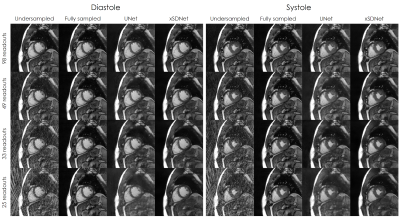
Figure 2: Results of the different image reconstructions for one exemplary subject in diastolic and systolic view.
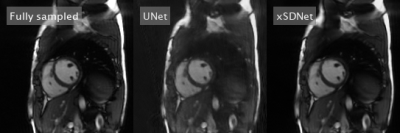
Figure 3: Dynamic view: Comparison of fully sampled reference with UNet- and xSDNet- reconstruction of 33 readouts per frame for another exemplary subject.
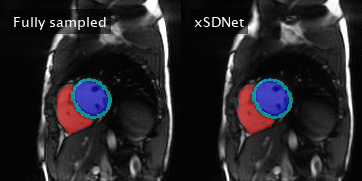
Figure 4: Left: Fully sampled image series with reference segmentation
masks superimposed. The latter correspond to the reference labels
obtained by
applying Bai’s model on the subadjacent images. Right: Image series and
segmentation masks as obtained by xSDNet applied to frames acquired with
33 projections each.
Table 1: Left: Mean and SD of the ratings performed by two experienced
radiologists (grey/white) on a 5-point Likert scale for the following
categories: Spatial resolution (1-poor to 5-high), artifact level
(1-severe to 5-no), contrast between myocardium and blood (1-poor to
5-high), signal-to-noise-ratio (1-low to 5-high), overall image
impression (1-poor to 5-excellent). On the right hand side, the Dice
coefficient between the segmentations obtained by xSDNet applied to
undersampled data and the reference masks as obtained by Bai's model on
fully sampled data are given.
DOI: https://doi.org/10.58530/2022/0016