S25
Differential Diagnosis of uterine leiomyoma pathological subtypes: A Conditional Inference Tree Model Based on MRI-T2WI
Chao Wei1 and Jiangning Dong1
1Anhui provincial tumor hosptial, Hefei, China
1Anhui provincial tumor hosptial, Hefei, China
Synopsis
The establishment and verification of conditional inference tree model based on based on the MRI-T2WI radiomics has high clinical value in the identification of three common pathological subtypes of uterine leiomyoma.
Purpose
To explore the value of the conditional inference tree model based on the MRI-T2WI radiomics in the identification of common pathological subtypes of uterine leiomyoma.Materials and Methods
Retrospective analysis was performed on 65 uterine leiomyomas confirmed by pathology with conventional MRI and DWI, including normal type(21 cases),cellular type(23 cases),degenerate type(21 cases)All lesions were randomly divided into the training group and the verification group (approx. 7:3). ITK-Snap software was used to delineate the region of interest on every section of T2WI along the lesion margin. A.K. software was used to extract the radiomics features at region of interest. Feature screening was performed by Pearson correlation analysis and random forest function. The selected features were used to construct a conditional inference tree model "ctree". The ROC curve of the validation group was used to evaluate the differential diagnosis capability of the model.Results
720 radiomics features were extracted from each lesion, and 12 features were obtained after filtering and screening to establish the conditional inference tree model. The area under ROC curve of the normal type, cellular type and degenerate type in the traning group was 0.97,0.82 and 0.91, respectively; the sensitivity was 100%,70.6% and 80%, respectively; the specificity was 93.8%,90% and 90.6%, respectively. In the verification group, the AUC of the above three types was 0.92,0.73 and 0.78,the sensitivity was85.3%,50% and 66.7%, respectively; the specificity was 100%,75% and 75%,respectively.Conclusion
The establishment and verification of conditional inference tree model based on based on the MRI-T2WI radiomics has high clinical value in the identification of three common pathological subtypes of uterine leiomyoma.Acknowledgements
We thank Shao-feng Duan from GE Healthcare China for technical assistance. This work was supported by the Anhui Provincial Natural Science Foundation for Youths, China (1908085QH364),References
- Wang Y, Wang Z B, Xu Y H. Efficacy, Efficiency, and Safety of Magnetic Resonance-Guided High-Intensity Focused Ultrasound for Ablation of Uterine Fibroids: Comparison with Ultrasound-Guided Method[J]. Korean J Radiol. 2018, 19(4): 724-732.
- Schwartz L B, Zawin M, Carcangiu M L, et al. Does pelvic magnetic resonance imaging differentiate among the histologic subtypes of uterine leiomyomata?[J]. Fertility and sterility. 1998, 70(3): 580-587.
- Funaki K, Fukunishi H, Funaki T, et al. Magnetic resonance-guided focused ultrasound surgery for uterine fibroids: relationship between the therapeutic effects and signal intensity of preexisting T2-weighted magnetic resonance images[J]. Am J Obstet Gynecol. 2007, 196(2): 181-184
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis[J]. Eur J Cancer. 2012, 48(4): 441-446.
- Lakhman Y, Veeraraghavan H, Chaim J, et al. Differentiation of Uterine Leiomyosarcoma from Atypical Leiomyoma: Diagnostic Accuracy of Qualitative MR Imaging Features and Feasibility of Texture Analysis[J]. European Radiology. 2017, 27(7).
- Hricak H, Tscholakoff D, Heinrichs L, et al. Uterine leiomyomas: correlation of MR, histopathologic findings, and symptoms[J]. Radiology. 1986, 158(2): 385-391
- Takahashi M, Kozawa E, Tanisaka M, et al. Utility of histogram analysis of apparent diffusion coefficient maps obtained using 3.0T MRI for distinguishing uterine carcinosarcoma from endometrial carcinoma[J]. J Magn Reson Imaging. 2016, 43(6): 1301-1307.
- Sumi A, Terasaki H, Sanada S, et al. Assessment of MR Imaging as a Tool to Differentiate between the Major Histological Types of Uterine Sarcomas[J]. Magn Reson Med Sci. 2015, 14(4): 295-304.
- Lakhman Y, Veeraraghavan H, Chaim J, et al. Differentiation of Uterine Leiomyosarcoma from Atypical Leiomyoma: Diagnostic Accuracy of Qualitative MR Imaging Features and Feasibility of Texture Analysis[J]. European Radiology. 2017, 27(7): 2903-2915.
- Nakagawa M, Nakaura T, Namimoto T, et al. Machine Learning to Differentiate T2-Weighted Hyperintense Uterine Leiomyomas from Uterine Sarcomas by Utilizing Multiparametric Magnetic Resonance Quantitative Imaging Features[J]. Acad Radiol. 2019, 26(10): 1390-1399.
Figures
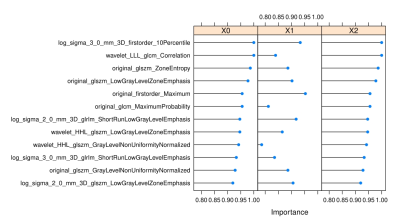
Distribution of features sorted by conditional reasoning tree model in three groups.The y-axis represents the feature name, the x-axis represents the relative weight of the feature, the line with blue dot in the middle represents the importance of each feature in predicting this subtype RCC, and the longer the line is, the more important the feature is.
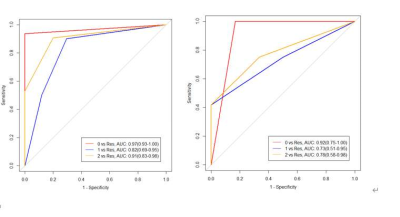
The ROC curves of normal type, cellular type and degenerate type were identified by conditional reasoning tree model. The left picture was training group, The right picture was validation group. 0 is normal type, 1 is cellular type, 2 was degenerate type, res was the remaining two groups.