Application of MRI to DBS Patients
1University of Minnesota, Minneapolis, MN, United States
Synopsis
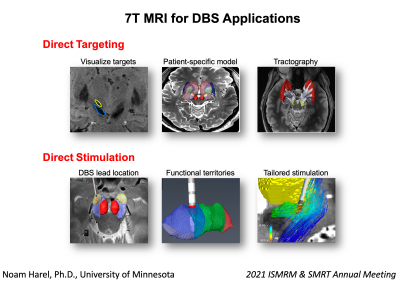
Deep brain stimulation (DBS) can be highly effective at improving symptoms and enhancing the patient’s quality of life. However, clinical outcomes can vary greatly within- and across-studies. Much of this variability can likely be accounted for by differences in the stimulating lead location. High resolution 7T MRI data provides a unique opportunity to create an accurate, patient-specific, 3D anatomical model for visualization of the target structures. Accurate lead location, in respect to the anatomical borders of the intended target structure and its functional domains, is a crucial information for an efficient programming for patient optimal benefits.
Acknowledgements
National Institution of Health; P50 NS098753, R01 NS081118, R01 NS113746References
1. Starr, P.A., et al., Magnetic resonance imaging-based stereotactic localization of the globus pallidus and subthalamic nucleus. Neurosurgery, 1999. 44(2): p. 303-13; discussion 313-4.
2. Rolston, J.D., et al., An unexpectedly high rate of revisions and removals in deep brain stimulation surgery: Analysis of multiple databases. Parkinsonism Relat Disord, 2016. 33: p. 72-77.
3. Abosch, A., et al., An assessment of current brain targets for deep brain stimulation surgery with susceptibility-weighted imaging at 7 tesla. Neurosurgery, 2010. 67(6): p. 1745-56; discussion 1756.
4. Forstmann, B.U., et al., Multi-modal ultra-high resolution structural 7-Tesla MRI data repository. Sci Data, 2014. 1: p. 140050.
5. Duchin, Y., et al., Patient-specific anatomical model for deep brain stimulation based on 7 Tesla MRI. PLoS One, 2018. 13(8): p. e0201469.
6. Plantinga, B.R., et al., Individualized parcellation of the subthalamic nucleus in patients with Parkinson's disease with 7T MRI. Neuroimage, 2018. 168: p. 403-411.
7. Lenglet, C., et al., Comprehensive in vivo mapping of the human basal ganglia and thalamic connectome in individuals using 7T MRI. PLoS One, 2012. 7(1): p. e29153.
8. Keuken, M.C., et al., Quantifying inter-individual anatomical variability in the subcortex using 7 T structural MRI. Neuroimage, 2014. 94: p. 40-46.
9. Patriat, R., et al., Individualized tractography-based parcellation of the globus pallidus pars interna using 7T MRI in movement disorder patients prior to DBS surgery. Neuroimage, 2018. 178: p. 198-209.
10. Behrens, T.E., et al., Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat Neurosci, 2003. 6(7): p. 750-7.
11. Shamir, R.R., et al., Microelectrode Recordings Validate the Clinical Visualization of Subthalamic-Nucleus Based on 7T Magnetic Resonance Imaging and Machine Learning for Deep Brain Stimulation Surgery. Neurosurgery, 2018.
12. Aman, J.E., et al., Directional deep brain stimulation leads reveal spatially distinct oscillatory activity in the globus pallidus internus of Parkinson's disease patients. Neurobiol Dis, 2020. 139: p. 104819.
13. Schrock, L.E., et al., 7T MRI and Computational Modeling Supports a Critical Role of Lead Location in Determining Outcomes for Deep Brain Stimulation: A Case Report. Front Hum Neurosci, 2021. 15: p. 631778.
14. Kim, J., et al., Automatic localization of the subthalamic nucleus on patient-specific clinical MRI by incorporating 7 T MRI and machine learning: Application in deep brain stimulation. Hum Brain Mapp, 2019. 40(2): p. 679-698.
15. Solomon, O., et al., Deep-learning based fully automatic segmentation of the globus pallidus interna and externa using ultra-high 7 Tesla MRI. Hum Brain Mapp, 2021.