4300
GPU-accelerated diffusion MRI tractography in DIPY1Psychology, University of Washington, Seattle, WA, United States, 2NVIDIA, Menlo Park, CA, United States, 3University of California, San Francisco, San Francisco, CA, United States, 4University of Washington, Seattle, WA, United States, 5Indiana University, Bloomington, IN, United States
Synopsis
Tractography based on diffusion-weighted MRI provides non-invasive in vivo estimates of trajectories of long-range brain connections. These estimates are important in research that measures individual differences in brain connections and in clinical use-cases. But the computational demands of tractography present a barrier to progress. Here, we present a GPU-based tractography implementation that accelerates tractography algorithms implemented as part of the Diffusion Imaging in Python (DIPY) project. This implementation speeds up tractography by at least a factor of ~200X, providing tractographies that closely match CPU-based solutions. These speedups enable applications of tractography in clinical data, and in very large datasets.
Introduction
The white matter of the brain contains the axons of long-range connections between distant brain regions. The integration and coordination of brain activity through these connections are important for information processing and for brain health. Diffusion-weighted MRI (dMRI) and computational tractography provide non-invasive in vivo estimates of the trajectories of these brain connections. This is done by estimating the directions of diffusion in every voxel of measurement, and by propagating streamlines through the volume based on the peaks in the distribution of diffusion directions. The resulting tractograms are important in research that uses dMRI to measure individual differences in brain connections. They are also important in clinical use-cases, where invasive procedures are guided by these estimates, aiming to avoid disconnections of crucial brain pathways. However, a major barrier to progress in the use of these methods is that tractography can be a computationally-intensive processing step. Even when highly-efficient CPU-based implementations are provided in open-source software, these can take hours to process millions of streamlines in an individual brain. By taking advantage of the massively parallel architecture of graphical processing units (GPU) and the implementations of very fast basic computational operations, GPU-based computing can be used to accelerate many scientific use-cases.Methods
DIPY (Diffusion Imaging in Python; https://dipy.org) is an open-source software library that implements many methods in computational neuroanatomy1. Relying on the DIPY implementation of residual bootstrap tractography2, we implemented a multi-GPU parallelizable version constructed on NVIDIA’s CUDA application programming interface (API). The API of the GPU version is compatible with the one implemented in DIPY, enabling direct comparisons and interoperability. A docker container of the software makes the installation and use of the software straightforward. The software is available at https://github.com/dipy/GPUStreamlines. Experiments to profile the performance of the algorithm were conducted using an AWS p3.16xlarge instance with 8 NVIDIA Tesla V100 Graphical Processing Units and 488 GB RAM. For comparison, CPU code was run on an AWS x1e.4xlarge with 488 GB RAM. We used two datasets, the first is a HARDI acquisition with 2x2x2 mm3 isotropic voxels, 150 b=1,000 s/mm2 volumes and 10 b0 volumes previously described3. The other dataset was a Super-Resolution Hybrid Diffusion Imaging (HYDI) dataset4, with an effective resolution of 0.625 mm3 isotropic voxels, b=500, 800, 1600, 2600 s/mm2, in 134 diffusion directions, and 8 b0 volumes, also previously described5. In both cases, 27 seeds were placed in each voxel in the white matter to initialize tracking.Results
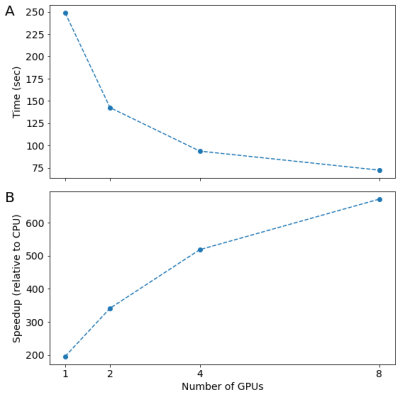
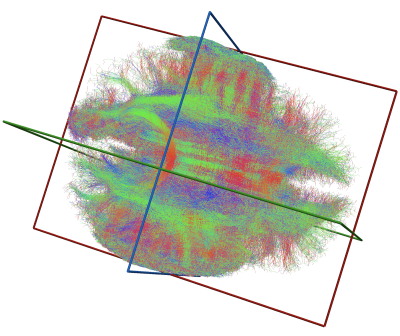
In the HARDI dataset, with the seeding approach used here, approximately 2.1M streamlines were generated. Using the CPU-based residual bootstrap tracking algorithm took approximately 13 hours. The GPU-accelerated implementation provides approximately 200-fold speedup with a single GPU, and up to 671-fold speedup with 8 GPUs run in parallel (Figure 1). In the HYDI dataset, the seeding approach used generated 150M streamlines (497GB). Tracking in this case with 8 GPU completed in just under 2 hours. A subset of the HYDI streamlines is shown in Figure 2.Discussion and Conclusion
A GPU-based implementation of residual bootstrap tractography provides orders of magnitude speedup, relative to the CPU-based version, while providing solutions that match CPU-based solutions very closely. This was demonstrated in standard and high-resolution measurements. Thus, this GPU-based implementation allows researchers to both (1) save time and money solving existing problem sizes and (2) solve new problems that are computationally intractable on CPU-only resources. Open-source software is provided, as well as a docker container that encapsulates the software, together with all of its dependencies available at docker.pkg.github.com/dipy/gpustreamlines/gpustreamlines.Acknowledgements
DIPY development is supported through grant 5R01EB027585-02 to Eleftherios Garyfallidis and Ariel RokemReferences
1. Garyfallidis, E., Brett, M., Amirbekian, B., Rokem, A., van der Walt, S., Descoteaux, M., Nimmo-Smith, I. & Dipy Contributors. Dipy, a library for the analysis of diffusion MRI data. Front. Neuroinform. 8, 8 (2014).
2. Berman, J. I., Chung, S., Mukherjee, P., Hess, C. P., Han, E. T. & Henry, R. G. Probabilistic streamline q-ball tractography using the residual bootstrap. Neuroimage 39, 215–222 (2008).
3. Rokem, A., Yeatman, J. D., Pestilli, F., Kay, K. N., Mezer, A., van der Walt, S. & Wandell, B. A. Evaluating the accuracy of diffusion MRI models in white matter. PLoS One 10, e0123272 (2015).
4. Garyfallidis, E., Nahla, E. & Wu, Y.-C. Superresolved HYDI dataset. (2019). doi:10.6084/m9.figshare.10266194.v15. Elsaid, N. M. H., Coupé, P. & Wu, Y.-C. Super-Resolution Hybrid Diffusion Imaging. in International Society for Magnetic Resonance in Medicine (ISMRM) at <http://archive.ismrm.org/2019/3349.html>
Figures