4248
Deep Learning-based MR-only Radiation Therapy Planning for Head&Neck and Pelvis
Florian Wiesinger1, Sandeep Kaushik1, Mathias Engström2, Mika Vogel1, Graeme McKinnon3, Maelene Lohezic1, Vanda Czipczer4, Bernadett Kolozsvári4, Borbála Deák-Karancsi4, Renáta Czabány4, Bence Gyalai4, Dorottya Hajnal4, Zsófia Karancsi4, Steven F. Petit5, Juan A. Hernandez Tamames5, Marta E. Capala5, Gerda M. Verduijn5, Jean-Paul Kleijnen5, Hazel Mccallum6, Ross Maxwell6, Jonathan J. Wyatt6, Rachel Pearson6, Katalin Hideghéty7, Emőke Borzasi7, Zsófia Együd7, Renáta Kószó7, Viktor Paczona7, Zoltán Végváry7, Suryanarayanan Kaushik3, Xinzeng Wang3, Cristina Cozzini1, and László Ruskó4
1GE Healthcare, Munich, Germany, 2GE Healthcare, Stockholm, Sweden, 3GE Healthcare, Waukesha, WI, United States, 4GE Healthcare, Budapest, Hungary, 5Erasmus MC, Rotterdam, Netherlands, 6Newcastle University, Newcastle, United Kingdom, 7University of Szeged, Szeged, Hungary
1GE Healthcare, Munich, Germany, 2GE Healthcare, Stockholm, Sweden, 3GE Healthcare, Waukesha, WI, United States, 4GE Healthcare, Budapest, Hungary, 5Erasmus MC, Rotterdam, Netherlands, 6Newcastle University, Newcastle, United Kingdom, 7University of Szeged, Szeged, Hungary
Synopsis
MR imaging offers unique advantages for Radiation Therapy Planning (RTP) via excellent soft-tissue contrast for the delineation of the tumor target volume and surrounding organs-at-risk (OARs). Remaining challenges include absent CT information (required for accurate dose calculation) and time-consuming manual tumor and OAR contouring. Here we describe the application of Deep Learning for MR-only RTP in terms of synthetic CT conversion and automated OAR delineation. Exemplary results are illustrated from an ongoing MR-only RTP study in head&neck and pelvis.
Introduction
MR imaging offers unique advantages for Radiation Therapy Planning (RTP) in terms of exquisite soft-tissue contrast for the delineation of the tumor target volume and surrounding organs-at-risk (OARs). Remaining challenges include absent CT information (required for accurate dose calculation) and time-consuming manual tumor target and OAR contouring1. Recently, new Deep-Learning (DL) based image processing methods have added powerful tools for image reconstruction, image translation and automated segmentation2. Here we take advantage of these developments and describe the application of Deep Learning for MR-only RTP in terms of synthetic CT conversion and automated OAR delineation. Exemplary results are illustrated from an ongoing MR-only RTP study in head&neck and pelvis.Methods
Synthetic CT conversion: RT patients need to be imaged with fixation devices. For head&neck application in particular, they need to wear a mask, that renders the imaging session very uncomfortable. For synthetic CT conversion we therefore developed a fast (1-2mins), large field-of-view (FOV=50cm), high-resolution (1.5mm for Head&Neck and 2.0mm for pelvis) in-phase Zero TE (ipZTE) acquisition method3 including DL-based image reconstruction4. The 3D ipZTE images provide uniform soft-tissue response with excellent bone depiction5 and hence are well-suited as input for DL-based synthetic CT conversion which was implemented in form of a modified 2D UNet architecture with continuous Hounsfield Unit [HU] assignment similar as described in reference6. The DL algorithm is designed to maintain the overall structural accuracy with an emphasized focus on accurate bone depiction. DL training was performed using pairs of registered ipZTE and CT patient images obtained from independent earlier studies (N=140 and 30 for head&neck and pelvis, respectively).Organ-At-Risk (OAR) segmentation: For OAR segmentation standard T2-weighted images (2D T2 PROPELLER for head&neck and 3D T2 CUBE for pelvis; each ~5mins scan time) are acquired including optional DL-based image reconstruction4. The two-step automated OAR segmentation starts with 2D DL-based organ localization followed by a 3D DL-based fine-segmentation within the cuboid bounding box7. For head&neck 20 OARs (i.e. eye L/R, lens L/R, optic nerve L/R, chiasm, brain, brainstem, pituitary gland, cochlea L/R, spinal cord, mandible, oral cavity, parotid gland L/R, submandibular gland L/R, larynx, and body outline) and for pelvis 5 OARs (i.e. femur head L/R, bladder, rectum, prostate, and body outline) are segmented. The DL network was trained based on manually contoured T2-weighted images obtained from independent studies (N=44 and 48 for head&neck and pelvis, respectively).
The DL-based methods for synthetic CT conversion and OAR segmentation were evaluated on a small set of patient cases obtained from an ongoing MR-only RTP study.
Results
Synthetic CT conversion: Figures 1 and 2 illustrate exemplary head&neck (patient #5) and pelvis (patient #8) ipZTE to synthetic CT image conversion results. The top two rows compare ipZTE images without (top) and with (2nd row) DL-based image reconstruction. Apparently, DL-based image reconstruction recovers a sharp ipZTE image behind an otherwise unacceptable noise background and thereby shortens the scan time to only ~1-2mins for high resolution (1.5-2mm) large FOV=50cm coverage. The bottom two rows compare the DL-derived synthetic CT (3rd row) relative to the true CT (4th row).Organ-At-Risk (OAR) segmentation: Figures 3 and 4 illustrate exemplary T2-based OAR segmentation results for head&neck (using 2D T2 PROPELLER, ~5mins) and pelvis (using 3D CUBE T2, ~5mins) for the same patients as mentioned above. The left subplots illustrate the input T2 images with standard (top) and DL-based (bottom) reconstruction. The automatically segmented OARs are displayed with color contours. The right subplots show semi-transparent 3D rendering of the segmented OARs.
Discussion
The preliminary results of this ongoing study demonstrate the technical feasibility of DL-based MR-only RTP in terms of synthetic CT conversion and OAR delineation for static head&neck and pelvis anatomies. DL-based MR to synthetic CT image translation eliminates the need for an extra planning CT (required for dose calculation) and thereby enables an MR-only workflow. On the other hand, DL-based OAR auto-contouring shortcuts the manual contouring (~4h per patient) resulting in significant time and cost savings. Using standardized MR imaging protocols in combination with DL-based MR image reconstruction also shortens the overall scan time which is specifically important for MR imaging in fixation using e.g. a thermoplastic head mask as it is usually the case for head&neck patients.Acknowledgements
This research is part of the Deep MR-only Radiation Therapy activity (project numbers: 19037, 20648) that has received funding from EIT Health. EIT Health is supported by the European Institute of Innovation and Technology (EIT), a body of the European Union and receives support from the European Union´s Horizon 2020 research and innovation program.References
- McGee et al (2016). MRI in radiation oncology: Underserved needs. MRM 75(1), 11-4.
- LeCun et al (2015). Deep learning. Nature 521(7553), 436-44.
- Engström et al (2020). In‐phase zero TE musculoskeletal imaging. MRM 83(1), 195-202.
- Lebel (2020). Performance characterization of a novel deep learning-based MR image reconstruction pipeline. arXiv preprint arXiv:2008.06559.
- Wiesinger et al (2018). Zero TE‐based pseudo‐CT image conversion in the head and its application in PET/MR attenuation correction and MR‐guided radiation therapy planning. MRM 80(4), 1440-1451.
- Kaushik et al (2018). Deep Learning based pseudo-CT computation and its application for PET/MR attenuation correction and MR-guided radiation therapy planning. ISMRM 2018: p.1253.
- Rusko et al (2020). Automated organ delineation in T2 head MRI using combined 2D and 3D convolutional neural networks. ESTRO 2020: PO-1709.
Figures
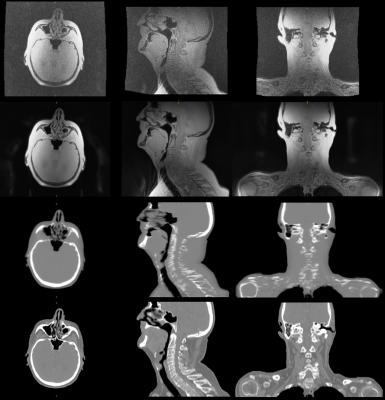
Figure 1: In-phase
ZTE without (top) and with 2xFOV extension plus DL image reconstruction (2nd
row) for head&neck, together with corresponding DL derived synthetic CT (3rd
row) and true CT (bottom row).
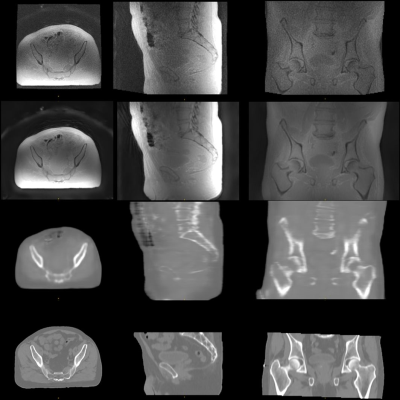
Figure 2: In-phase ZTE without (top) and with 2xFOV extension plus DL image reconstruction (2nd row) for pelvis, together with corresponding DL derived synthetic CT (3rd row) and true CT (bottom row).
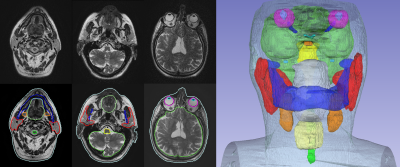
Figure 3: 2D T2 PROPELLER based
automated OAR segmentation in the head & neck (i.e. brain, brainstem, eyes,
lens, optic nerves, chiasm, pituitary gland, cochlea, parotid glands, mandible,
oral cavity, submandibular glands, larynx, spinal cord, and body contour).
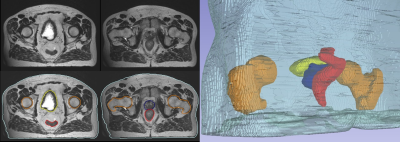
Figure 4: 3D T2 CUBE based
automated OAR segmentation in the pelvis (i.e. femurs, bladder, prostate, rectum,
and body contour).