4218
Fast T1 Measurement of Cortical Bone using 3D Ultrashort Echo Time Actual Flip Angle Imaging and Single Repetition Time Acquisition (UTE-AFI-STR)1Department of Radiology, UC San Diego, San Diego, CA, United States, 2Institute of Electrical Engineering, Chinese Academy of Sciences, Beijing, China, 3University of Chinese Academy of Sciences, Beijing, China
Synopsis
To improve the time efficiency for cortical bone T1 measurement on clinical scanners, we proposed a new method which fits the datasets of 3D ultrashort echo time actual flip angle imaging (UTE-AFI) and UTE with a single TR (UTE-STR) simultaneously (UTE-AFI-STR). The results show that the UTE-AFI-STR can measure the T1 of cortical bone accurately and more efficiently than the UTE-AFI-VTR method.
Introduction
Ultrashort echo time (UTE) sequences have been used for cortical bone water imaging and quantification in order to acquire useful magnetic resonance (MR) signals1. The T1 is a useful biomarker for the cortical bone porosity and age related deterioration assessment and is also a sensitive biomarker for monitoring temperature change in cortical bone2,3. Recently, we proposed a 3D UTE based actual flip angle imaging (AFI)-variable TR (VTR) (UTE-AFI-VTR) method for accurate cortical bone T1 measurement4. The UTE-AFI-VTR method can achieve accurate measurement of the T1 of cortical bone without degradation from the inefficient RF excitation of clinical scanners4. However, the scan time of the UTE-AFI-VTR sequence was relatively long for clinical practice because the UTE-VTR dataset with multiple TRs were required. To improve the time efficiency for accurate cortical bone T1 measurement, we proposed a generalized optimization framework which simultaneously fits the UTE-AFI and the UTE with a single TR (UTE-STR) data sets to estimate T1. This method would significantly reduce the total scan time for T1 measurement.Methods
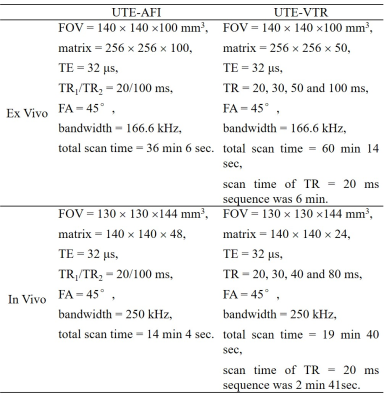
The features of the 3D UTE-AFI and UTE-VTR sequences are shown in Figure 14. A short rectangular RF pulse (duration 150 μs) is used for signal excitation and this is followed by a spiral trajectory data acquisition with 3D conical view ordering in these two sequences. For ultrashort T2 tissue excitation, the steady-state signals in TR1 and TR2 of the 3D UTE-AFI sequence conform to the signal models described by Eqs. [1] and [2] in reference 4. The signal detected by the 3D UTE sequence is described by Eq. [9] in reference 4. In addition, since the equilibrium magnetization M0 and transverse magnetization mapping function generated by the RF pulse Fxy are independent of TR, they can be combined into a single unknown parameter (e.g., κ). As a result, when the RF pulse and flip angle (FA) used in the AFI and VTR sequences are identical, there are only three unknown parameters (i.e., κ, T1, Fz) in these signal models to fit. Therefore, we can use the optimization framework below to simultaneously fit (sf) UTE-AFI and UTE-VTR (sf-UTE-AFI-VTR) data:$$\left[\kappa~T_{1}~F_{z}\right]=\text{arg}\min\limits_{\kappa,T_{1},F_{z}}\left\{\sum_{i=1}^2[I_{AFI,i}-S_{AFI,i}]^2+\sum_{j=1}^N[I_{VTR,j}-S_{VTR,j}]^2\right\}~~~~~~~[1]$$
where $$$\text{arg}\min\limits_{x}\left(y(x)\right)$$$ means finding a $$$x$$$ value which can make $$$y(x)$$$ attain its minimum value. $$$I_{AFI,i}$$$ ($$$i$$$ = 1 and 2) and $$$I_{VTR,j}$$$ ($$$j$$$ = 1,2,..., N) are the sampled signal intensities from the UTE-AFI and UTE-VTR sequences respectively. $$$S_{AFI,i}$$$ and $$$S_{VTR,j}$$$ are the signal models of the UTE-AFI and UTE-VTR sequences respectively4. N is the total number of TRs. $$$\kappa=M_{0}F_{xy}\left(\alpha,\tau,T_{2}\right)$$$. This framework simultaneously yields a solution for the three unknown parameters: κ, T1, and Fz. Since there are only three unknown parameters in Eq. [1], a single regular UTE acquisition together with the other two UTE-AFI acquisitions is sufficient to estimate them using the optimization framework. To save scan time, we therefore propose a UTE-AFI-STR method (i.e., a combination of the UTE-AFI and UTE-STR methods) to provide accurate measurement of T1.
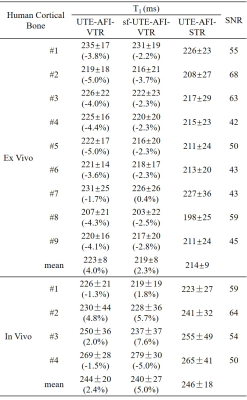
Numerical simulations, ex vivo and in vivo human cortical bone experiments were conducted to demonstrate the accuracy of the proposed fast 3D UTE-AFI-STR method. In simulations, T1 maps generated with the UTE-AFI-VTR, sf-UTE-AFI-VTR, and UTE-AFI-STR methods were compared at different SNR levels and the error ratios were calculated. The UTE-AFI and UTE-VTR sequences were performed on nine human cortical bone samples (aged from 38 to 95 years, 5 females) and four healthy volunteers (35±16 years, 3 males) and their T1 maps were calculated with the aforementioned three methods. The T1 value difference errors between the UTE-AFI-STR and the UTE-AFI-VTR/sf-UTE-AFI-VTR methods were calculated as well. The parameters used in the simulations were the same as in vivo experiments and the sequence parameters of ex vivo and in vivo experiments are shown in Table 1. The total scan times of the UTE-AFI-STR method were about 54 minutes and 17 minutes less than the UTE-AFI-VTR/sf-UTE-AFI-VTR methods in the ex vivo and in vivo experiments, respectively.
Results and Discussion
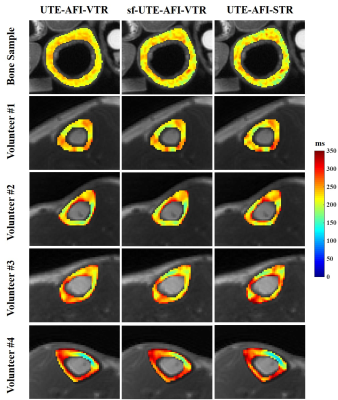
As shown in Figure 2, when the signal to noise ratio (SNR) of UTE-STR image was higher than 40, the T1 value derived from UTE-AFI-STR method was close to the standard T1 (i.e., 220 ms) (error ratios ranged from -5% to 5%). This observation shows that the fast UTE-AFI-STR method is accurate for T1 measurement when the SNR of the UTE-STR image is higher than 40.In Figure 3, the T1 maps of ex vivo and in vivo cortical bones obtained with three methods looked similar. In Table 2, the T1 value difference ratios between range between -5.0% and 0.4%, and -5.0% to 7.6% for ex vivo and in vivo measurements, respectively. The average cortical bone T1 for the four volunteers calculated using the UTE-AFI-STR method was 246 ms, which is similar to previously reported values5-7. These observations demonstrate the accuracy and good agreement of the T1 measurement of the UTE-AFI-STR method as compared with the other two methods which require much more scan time.
Conclusion
The proposed 3D UTE-AFI-STR method provides accurate T1 mapping of cortical bone with improved time efficiency compared with the UTE-AFI-VTR/sf-UTE-AFI-VTR methods.Acknowledgements
The authors acknowledge grant support from GE Healthcare, NIH (R01AR068987, R01NS092650, and R21AR075851) and scholarship support from the Joint Ph.D. Training Program of the University of Chinese Academy of Sciences (UCAS).References
1. Du J, Bydder GM. Qualitative and quantitative ultrashort-TE MRI of cortical bone. NMR Biomed. 2013;26:489–506.
2. Akbari A, Abbasi-Rad S, Rad HS. T1 correlates age: A short-TE MR relaxometry study in vivo on human cortical bone free water at 1.5T. Bone 2016;83:17–22.
3. Han M, Rieke V, Scott SJ, et al. Quantifying temperature-dependent T1 changes in cortical bone using ultrashort echo-time MRI. Magn. Reson. Med. 2015;74:1548–1555.
4. Ma YJ, Lu X, Carl M, et al. Accurate T1 mapping of short T2 tissues using a three-dimensional ultrashort echo time cones actual flip angle imaging-variable repetition time (3D UTE-Cones AFI-VTR) method. Magn. Reson. Med. 2018;80:598–608.
5. Yarnykh VL. Actual flip-angle imaging in the pulsed steady state: A method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn. Reson. Med. 2007;57:192–200.
6. Du J, Carl M, Bydder M, Takahashi A, Chung CB, Bydder GM. Qualitative and quantitative ultrashort echo time (UTE) imaging of cortical bone. J. Magn. Reson. 2010;207:304–311.
7. Chen J, Chang EY, Carl M, et al. Measurement of bound and pore water T1 relaxation times in cortical bone using three-dimensional ultrashort echo time cones sequences. Magn. Reson. Med. 2017;77:2136–2145.
Figures