4120
Non-invasive Gleason Score Classification with VERDICT-MRI1Centre for Medical Image Computing, Department of Computer Science, University College London, London, United Kingdom, 2Centre for Medical Imaging, University College London, London, United Kingdom
Synopsis
This study proposes non-invasive Gleason Score (GS) classification for prostate cancer with VERDICT-MRI using convolutional neural networks (CNNs). We evaluate GS classification using parametric maps from the VERDICT prostate model with compensated relaxation. We classify lesions using two CNN architectures: DenseNet and SE-ResNet. Results show that VERDICT achieves high GS classification performance using SE-ResNet with all parametric maps as input. Also in comparison with published GS classification multi-parametric MRI studies, VERDICT maps achieve higher metrics.
Introduction
Prostate cancer (PCa) diagnosis requires transrectal or transperineal biopsy, which is invasive and uncomfortable. Early diagnosis and treatment planning are important to reduce the mortality rate of PCa. Gleason score (GS) is the standard for measuring PCa aggressiveness and is obtained after histological inspection. Accurate GS is required to ensure prompt and correct PCa treatment.Multi-parametric MRI (mp-MRI) is the standard care for PCa diagnosis in the UK, however the specificity still requires improvement1. Vascular, Extracellular, and Restricted Diffusion for Cytometry in Tumours (VERDICT) is an advanced microstructural imaging technique for PCa characterisation1,2. VERDICT has demonstrated clear feasibility and repeatability in the clinical setting with the Intracellular (IC) volume fraction (fic) demonstrating capability to discriminate GS 3+4 from GS 3+3 10.
Machine learning methods have been employed for PCa classification4,5,6,7,8 using mp-MRI. A recent survey9 reports that most machine learning PCa applications use mp-MRI to solve binary classification (no cancer vs significant cancer) rather than GS. Five-point GS classification remains a challenging task with non-invasive imaging i.e. mp-MRI with apparent diffusion coefficient (ADC), high b-value (HBVAL) diffusion-weighted imaging (DWI), T2W images and dynamic contrast-enhanced (DCE). The best-performing method in a prostate imaging challenge (Prostate-X2) achieved only a weak weighted kappa value (metric for measuring agreement between estimated GS and ground truth obtained by pathology) 4. Methods using convolutional neural networks (CNNs) for GS classification with mp-MRI have not yet achieved stable results 6,7,8.
In this work, we evaluate GS classification using VERDICT maps generated with a recently proposed model that incorporates compartment specific T1/T2 relaxation 3. We classify cancer lesions into a five-point GS using two CNNs: DenseNet 12 and SE-ResNet 13.
Methods
- Data Acquisition
- Data Analysis
$$S(b,TE,TR)=S_0 (1 - e^\frac{-TR}{T1})[f_{vasc} e^\frac{-TE}{T2_{vasc/ees}}S_{vasc}(D_{vasc}= 8 m^2/ms,b)\\+ f_{ic} e^\frac{-TE}{T2_{ic}}S_{ic}(D_{ic}= 2 m^2/ms,R,b) + f_{ees} e^\frac{-TE}{T2_{vasc/ees}}S_{ees}(D_{ees},b)]$$ (Eq.1)
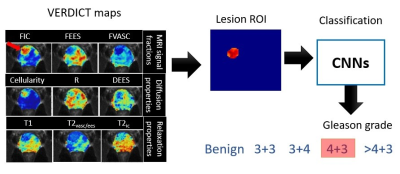
where $$$ fvasc+ fic+ fees=1 $$$. Figure 1 demonstrates the VERDICT maps: vascular (VASC), intracellular (IC), extracellular-extravascular (EES) volume fractions (fvasc, fic, fees), cellularity, cell radius(R), diffusion coefficient (DEES), T1 relaxation, slow vascular T2 compartment (T2vasc/ees), and fast T2 by intracellular (T2ic).
Model fitting: We implement Multilayer Perceptron (MLP) regression-based model fitting 2, 3. We trained three hidden layers MLP consisting of 150 hidden units with a nonlinearity function rectified linear unit (ReLU) and a final regression layer with synthetic DW-MRI signal using Eq.1. The trained MLP then predicts and generates VERDICT maps.
Classification: We perform classification on predefined lesion ROIs with VERDICT maps using CNNs. We consider five classes: Benign (34%), GS 3+3 (14%), GS 3+4 (28%), GS 4+3 (17%), GS >4+3 (7%). For the data imbalance, we use 5-fold cross-validation (test size is 20%) with stratified randomized folds to preserve the percentage of samples for each class. For training, we crop the images selecting the ROIs’ bounding box and apply data augmentation using rigid transformation. We train the networks for 30 epochs with Adam optimization at a learning rate of 10-5.
CNNs: We evaluate two CNNs. We consider DenseNet, a CNN that utilises dense connection between layers to connect all layers 12. DenseNet requires less computation while maintains high performance12. The second model is SE-Res-Net13, ResNet with embedded “Squeeze-and-Excitation (SE)" block that adaptively recalibrates feature response. SE blocks intrinsically introduce dynamics conditioned on the input, helping to boost feature discriminability. In SE-ResNet, SE was added before summation with the identity branch13.
Baseline: We compare our results with previous studies on five-point GS classification and mp-MRI data from ProstateX-2 challenge4 with pre-trained Inception-V35 and VGG networks7. Other studies with mp-MRI implement joint lesion segmentation and GS classification with U-Net6 and attention-based U-Net8.
Results
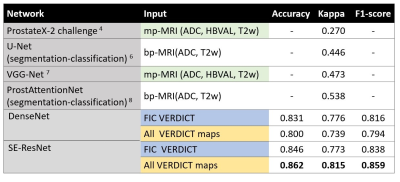
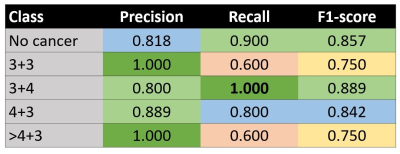
Fig 2. shows the overall performance metrics of GS classification using VERDICT with DenseNet and SE-ResNet. Compared to literature, VERDICT provides much higher kappa scores. We also compare using fic and all VERDICT maps (each map is treated as channel) as input for classification, where they perform equally well for both networks. SE-ResNet with all VERDICT maps achieves the highest GS discrimination.Fig 3. shows the GS classification using SE-ResNet and all VERDICT maps for each GS class.
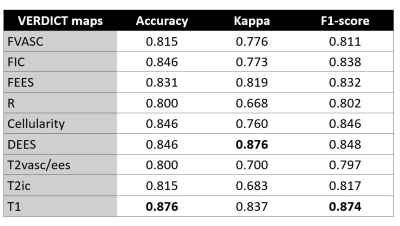
Fig 4. reports the performance metrics of GS classification by SE-ResNet using as input different VERDICT maps. We can see that other maps, such as DEES, cellularity and T1 perform well.
Discussion and Conclusion
This work explores the potential of VERDICT-MRI for GS classification with CNNs. Our results show that VERDICT with two different CNNs yields a high kappa score; substantial improvement (44%) over those found in literature with mp-MRI. Future work will include direct mp-MRI comparison with VERDICT on the same patients and a larger cohort.Acknowledgements
This project received funding from Engineering and Physical Sciences Research Council grand (EPSRC EP/N021967/1); INNOVATE trial; UKRI Future Leaders Fellowship MR/T020296/1 supports MP; the National Institute for Health Research (NIHR) University College London Hospitals (UCLH) Biomedical Research Centre.References
1.Panagiotaki, Eletheria, et al. "Noninvasive quantification of solid tumor microstructure using VERDICT MRI." Cancer research 74.7 (2014): 1902-1912.
2. Panagiotaki, Eleftheria, et al. "Microstructural characterization of normal and malignant human prostate tissue with vascular, extracellular, and restricted diffusion for cytometry in tumours magnetic resonance imaging." Investigative radiology 50.4 (2015): 218-227.
3. Palombo, Marco, et al. "Relaxed-VERDICT: decoupling relaxation and diffusion for comprehensive microstructure characterization of prostate cancer." Proc of International Society for Magnetic Resonance in Medicine (ISMRM) (2020).
4. Armato, Samuel G., et al. "PROSTATEx Challenges for computerized classification of prostate lesions from multiparametric magnetic resonance images." Journal of Medical Imaging 5.4 (2018): 044501.
5. Abraham, Bejoy, and Madhu S. Nair. "Computer-aided classification of prostate cancer grade groups from MRI images using texture features and stacked sparse autoencoder." Computerized Medical Imaging and Graphics 69 (2018): 60-68.
6. De Vente, Coen, et al. "Deep Learning Regression for Prostate Cancer Detection and Grading in Bi-parametric MRI." IEEE Transactions on Biomedical Engineering (2020).
7. Abraham, Bejoy, and Madhu S. Nair. "Automated grading of prostate cancer using convolutional neural network and ordinal class classifier." Informatics in Medicine Unlocked 17 (2019): 100256.
8. Duran, Audrey, Pierre-Marc Jodoin, and Carole Lartizien. "Prostate Cancer Semantic Segmentation by Gleason Score Group in mp-MRI with Self Attention Model on the Peripheral Zone." Medical Imaging with Deep Learning. 2020.
9. Arif, Muhammad, et al. "Automated classification of significant prostate cancer on MRI: a systematic review on the performance of machine learning applications." Cancers 12.6 (2020): 1606.
10. Johnston, Edward W., et al. "VERDICT MRI for prostate cancer: intracellular volume fraction versus apparent diffusion coefficient." Radiology 291.2 (2019): 391-397.
11. Johnston, Edward, et al. "INNOVATE: a prospective cohort study combining serum and urinary biomarkers with novel diffusion-weighted magnetic resonance imaging for the prediction and characterization of prostate cancer." BMC cancer 16.1 (2016): 816.
12. Huang, G., Liu, Z., Van Der Maaten, L., & Weinberger, K. Q. “Densely connected convolutional networks.” IEEE CVPR (2017): 4700-4708.
13. Hu, J., Shen, L., & Sun, G. "Squeeze-and-excitation networks.” IEEE CVPR (2018):7132-7141.
14. Panagiotaki E, Ianus A, Johnston E, Chan R, Stevens N, Atkinson D et al. “Optimised VERDICT MRI protocol for prostate cancer characterisation.” 23rd Scientific Meeting of the International Society for Magnetic Resonance in Medicine (2015).
Figures