4118
Sensitivity of radiomics to inter-reader variations in prostate cancer delineation on MRI should be considered to improve generalizability1Case Western Reserve University, Cleveland, OH, United States, 2University Hospitals Cleveland Medical Center, Cleveland, OH, United States, 3Cleveland Clinic, Cleveland, OH, United States
Synopsis
Radiomic approaches for prostate cancer risk stratification largely depend on radiologist delineation of prostate cancer regions of interest (ROI) on MRI. In this study, we acquired multi-reader delineations of ROIs, derived radiomic features within the ROIs trained and evaluated machine learning classifiers. We observed that variation in delineations did not affect the classification performance within a cohort but it did affect when evaluated on an independent validation set. We observed that a more conservative approach in delineations may ensure better generalizability and classification performance of machine learning models.
Introduction
Radiomics based predictive models using prostate MRI have been previously shown to enable better characterization of prostate cancer (PCa) and improved risk stratification 1–3. A large majority of these methods rely on manual delineation of prostate cancer (PCa) region of interest (ROI) on MRI by radiologist which may be influenced by inter-reader variations. The effect of these variations on performance and reliability of PCa predictive models has not been explored before. The purpose of our study is to investigate if radiomics derived from PCa ROIs delineated by multiple radiologists (N=3) on bi-parametric MRI (bpMRI: including T2W and ADC) significantly affect the performance of machine learning classifiers in identifying biopsy proven clinically significant PCa.Methods
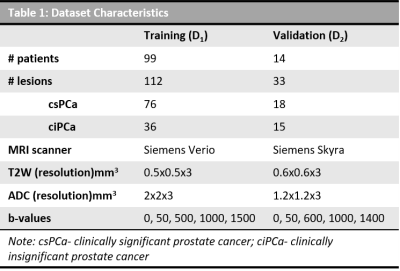
A publicly available dataset (D1) consisting of 99 patients4 with access to prostate 3T MRI scans, centroid location of PCa lesions and corresponding Gleason Grade Group (GGG) from targeted biopsy were included in the study. Patients with GGG=1 were considered to be with clinically insignificant PCa (ciPCa) and those with GGG>1 were with clinically significant PCa (csPCa). T2W MRI, ADC maps and the lesion location were provided to 3 experienced (>=7years) GU radiologists (R1, R2 & R3) for PCa delineation using 3D Slicer software. They were allowed to delineate as many MRI slices in which they considered the lesion to be visible. The overlap between inter-reader ROIs was evaluated in terms of dice similarity coefficient (DSC). T2W MRI intensities were standardized5 and radiomic texture features including 1st and 2nd order statistics, Haralick, Gabor, CoLlAGe6 and Laws were extracted within each of the 3 sets of ROIs on T2W and ADC. Features that showed significant differences between ciPCa and csPCa (Wilcoxon rank-sum test, p<0.05) were identified and used to train 3 logistic regression machine learning classifiers in conjunction with mrMR feature selection7 to predict csPCa, within a 3-fold, 150 run cross validation framework, for each set of ROIs. Partitioning of data within the folds was made consistent to ensure fair comparison. An independent validation dataset (D2) consisting of 14 PCa patients who underwent 3T MRI scan prior to radical prostatectomy was retrospectively acquired from an IRB approved, HIPAA compliant, anonymized cohort. PCa ROIs on bpMRI sequences for these patients were obtained from careful co-registration of whole mount specimens using a previously presented method8. The trained machine learning classifiers were evaluated on D2 and the performance was assessed in terms of area under the receiver operating characteristics curve (AUC). The agreement between predictions from classifiers trained using each of the three radiologist delineations was evaluated in terms of intra-class correlation coefficient (ICC(3,1)).Results
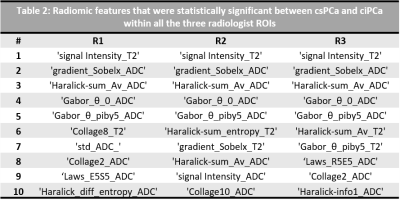
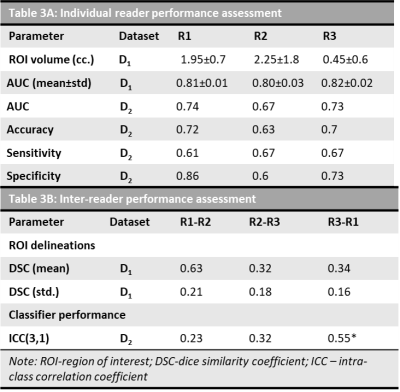
The dataset characteristics and imaging parameters are presented in Table 1. The mean volume of PCa ROIs delineated by R1, R2 and R3 are 1.95±0.7, 2.25±1.8, 0.45±0.6 respectively. The mean DSC for ROIs between pairs of readers (R1-R2, R2-R3, R3-R1) were 0.63, 0.32 and 0.34. Radiomic features that showed significant differences between csPCa and ciPCa across all the 3 sets of PCa ROIs included T2W mean intensities, gradient, Haralick and Gabor features from ADC maps (Table 2). Classifiers trained on D1 using PCa ROIs delineated by R1, R2 and R3 resulted in mean AUC of 0.81±0.18, 0.80±0.35 and 0.82±0.29 respectively in distinguishing csPCa and ciPCa. The AUCs on D2 in the same order were 0.74, 0.67 and 0.73. The ICC(3,1) values between classifier predictions from each pair of readers were 0.23, 0.32 and 0.55 respectively.Discussion
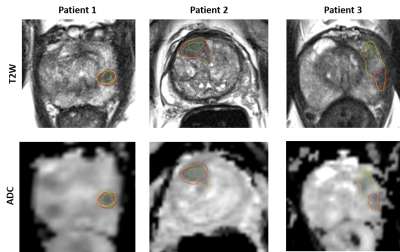
We observed that inter-reader variations in delineating PCa ROIs affected radiomic features that showed significant differences between csPCa and ciPCa. In Table 2, we observe Gabor features, that capture filter responses at multiple scales and orientations from ADC were consistent across all orientations. On T2W MRI, most texture features were not consistent across readers, which could potentially be due to its relatively higher resolution compared to ADC and texture features capturing underlying heterogeneity were dependent on the extent of ROI considered. The performance on D1 was good and relatively consistent in terms of AUC however when validated on D2, there were significant differences. The DSC measurements between R1-R2 was higher compared to the other pairs suggesting that R1 and R2 had much more consistent delineations. We notice that R2 tended to delineate larger ROIs compared to R1 and R3 (Table 3) and also illustrated in Figure 1. The larger delineations may result in inclusion of more noise from the ROI boundary which in turn affected classification performance on D2 as observed in Table 3. This is also reflected in terms of more consistent predictions on D2 between R1-R3 as evident from ICC(3,1) values. While previous studies9,10 have shown inclusion of peri-tumoral radiomics to improve prostate cancer risk stratification, a majority of those features were beyond 3mm from the boundary of the lesion. This suggests that under sampling the PCa ROI is beneficial for training reliable and generalizable machine learning classifiers.Conclusion
Inter-reader variations in delineating the prostate cancer regions of interest on MRI tend to affect radiomic features in distinguishing clinically significant and insignificant prostate cancer. Smaller regions of interest that under sample the lesion volume may result in more generalizable machine learning classifiers trained using radiomics.Acknowledgements
Research reported in this publication was supported by the National Cancer Institute under award numbers 1U24CA199374-01, R01CA202752-01A1R01CA208236-01A1R01CA216579-01A1R01CA220581-01A11U01CA239055-01 1U01CA248226-011U54CA254566-01National Heart, Lung and Blood Institute 1R01HL15127701A1National Institute for Biomedical Imaging and Bioengineering 1R43EB028736-01National Center for Research Resources under award number 1 C06 RR12463-01VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Servicethe Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program (W81XWH-19-1-0668)the Prostate Cancer Research Program (W81XWH-15-1-0558, W81XWH-20-1-0851)the Lung Cancer Research Program (W81XWH-18-1-0440, W81XWH-20-1-0595)the Peer Reviewed Cancer Research Program (W81XWH-18-1-0404)the Kidney Precision Medicine Project (KPMP) Glue Grantthe Ohio Third Frontier Technology Validation Fundthe Clinical and Translational Science Collaborative of Cleveland (UL1TR0002548) from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical ResearchThe Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University.
DoD Prostate Cancer Research Program Idea Development Award W81XWH-18-1-0524, Clinical and Translational Science Collaborative (CTSC) Cleveland Annual Pilot Award 2020 UL1TR002548
References
1. Lemaître G, Martí R, Freixenet J, Vilanova JC, Walker PM, Meriaudeau F. Computer-Aided Detection and diagnosis for prostate cancer based on mono and multi-parametric MRI: a review. Comput Biol Med. 2015 May;60:8–31. doi:10.1016/j.compbiomed.2015.02.009 PMID: 25747341
2. Viswanath S, Madabhushi A. Consensus embedding: theory, algorithms and application to segmentation and classification of biomedical data. BMC Bioinformatics. 2012;13:26. doi:10.1186/1471-2105-13-26 PMID: 22316103 PMCID: PMC3395843
3. Shiradkar R, Ghose S, Jambor I, Taimen P, Ettala O, Purysko AS, Madabhushi A. Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: Preliminary findings. J Magn Reson Imaging. 2018 May 7; doi:10.1002/jmri.26178 PMID: 29734484
4. Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H. Computer-Aided Detection of Prostate Cancer in MRI. IEEE Transactions on Medical Imaging. 2014 May;33(5):1083–1092. doi:10.1109/TMI.2014.2303821
5. Nyúl LG, Udupa JK. On standardizing the MR image intensity scale. Magn Reson Med. 1999 Dec;42(6):1072–1081. PMID: 10571928
6. Prasanna P, Tiwari P, Madabhushi A. Co-occurrence of Local Anisotropic Gradient Orientations (CoLlAGe): A new radiomics descriptor. Sci Rep. 2016 Nov 22;6:37241. doi:10.1038/srep37241 PMID: 27872484 PMCID: PMC5118705
7. Ding C, Peng H. MINIMUM REDUNDANCY FEATURE SELECTION FROM MICROARRAY GENE EXPRESSION DATA. Journal of Bioinformatics and Computational Biology. 2005 Apr;03(02):185–205. doi:10.1142/S0219720005001004
8. Li L, Pahwa S, Penzias G, Rusu M, Gollamudi J, Viswanath S, Madabhushi A. Co-Registration of ex vivo Surgical Histopathology and in vivo T2 weighted MRI of the Prostate via multi-scale spectral embedding representation. Sci Rep. 2017 Aug 18;7(1):8717. doi:10.1038/s41598-017-08969-w PMID: 28821786 PMCID: PMC5562695
9. Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, Pletcha D, Madabhushi A. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19. doi:10.1186/s13058-017-0846-1 PMID: 28521821 PMCID: PMC5437672
10. Algohary A, Shiradkar R, Pahwa S, Purysko A, Verma S, Moses D, Shnier R, Haynes A-M, Delprado W, Thompson J, Tirumani S, Mahran A, Rastinehad AR, Ponsky L, Stricker PD, Madabhushi A. Combination of Peri-Tumoral and Intra-Tumoral Radiomic Features on Bi-Parametric MRI Accurately Stratifies Prostate Cancer Risk: A Multi-Site Study. Cancers (Basel). 2020 Aug 6;12(8). doi:10.3390/cancers12082200 PMID: 32781640 PMCID: PMC7465024
Figures