4111
Machine learning challenge using uniform prostate MRI scans from 4 centers (PRORAD)1University of Turku, Turku, Finland, 2Turku University Hospital, Turku, Finland, 3Satakunta Central Hospital, Pori, Finland, 4Tampere University Hospital, Tampere, Finland, 5Helsinki University Hospital, Helsinki, Finland, 6Icahn School of Medicine at Mount Sinai, New York, NY, United States
Synopsis
PRORAD
is a series of machine learning challenges hosted at CodaLab which provide
access to prostate MRI data sets from 4 centers performed using a publicly available
IMPROD bpMRI acquisition protocol. The challenge is designed for purposes of developing,
validating and independent testing of various machine learning methods for prostate MRI.
INTRODUCTION
Magnetic Resonance Imaging (MRI) based radiomic and texture features hold great promise in the detection and characterization of prostate cancer diagnostics [1]. However, limitations in repeatability and reproducibility [2] contribute into reservations for adopting these techniques [3], suggesting image analysis competitions [4]. This prostate cancer detection and classification challenge is a one-time event, which follows general recommendations given in [5-6]. We developed a challenge to address situations where pre-calculated radiomic features from IMPROD bpMRI are available, but individual patient with suspected prostate cancer need to be classified to its correct Gleason Grade Group (GGG) [7] determined from Gleason score.MATERIALS & METHODS
We use MRI datasets collected using publicly available IMPROD bi-parametric Magnetic Resonance Imaging (bpMRI) protocol. The IMPROD bpMRI acquisition protocol consist of two separate T2-weighted imaging (1. axial; 2. sagittal) and three separate Diffusion Weighted Imaging acquisitions (1. b-values 0,100,200,300,500 s/mm2; 2) b-values 0,1500 s/mm2; 3) values 0,2000 s/mm2). The overall imaging time using 3 Tesla (T) scanners is approximately 13-17 minutes including shimming and calibration while the corresponding time at 1.5T is about 3 minutes longer. The detailed MRI protocol and importable MRI protocols are publicly available (http://petiv.utu.fi/improd and http://petiv.utu.fi/multiimprod).In this first part of a set of challenges (5 in total), radiomics and texture features were calculated from axial T2-weighted imaging and Diffusion Weighted Imaging acquisitions obtained using b-values of 0,100,200,300,500 s/mm2. Further details regarding IMPROD bpMRI protocol are provided in: PMID: 28165653, PMID: 30903647, PMID: 31158230, PMID: 31588660, PMID: 31750988. The publicly available feature vectors are anonymized and distributed in compliance with regulations for patient anonymity and data use.
Sample size:
- Site I: 3T MRI scans of 432 men
- Site II: 1.5T scans of 164 men
- Site III: 3T MRI scans of 58 men
- Site IV: 3T MRI scans of 58 men
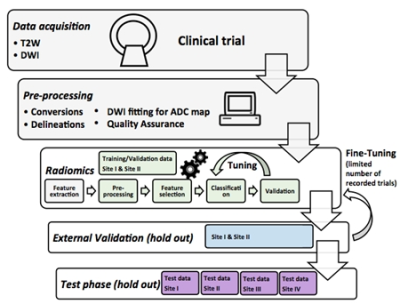
Details of challenge phases (Figure 1) are:
Training/Validation Phase
○ 12% data of Site I and 15% data of Site II
○ Evaluation is against same Gleason Grade Group (GGG) labels given to the user, for experimental use, unlimited number of submissions
○ Recorded performance scores are used in ranking at this phase
External Leaderboard Validation Phase
○ 12% data of Site I and 15% data of Site II
○ Validation against external unseen labels in hold-out set with a limited number of submissions.
○ Recorded performance scores are used in ranking at this phase
Test Phase
○ 76% data of Site I, 70% data of Site II, and 100% of Site III and IV data
○ One successful submission is required for participation to challenge
○ Scoring is allowed only for submissions with filled Test phase submission form e-mailed to aiforprostatemri@gmail.com. Please send another submission form (up to 5), if more submissions of significantly different approaches are desired.
○ Test phase is evaluated with binarized values, for this threshold.txt file is required to be submitted. The used threshold values are used to binarize the data as: value<threshold is 0 and value≥threshold is 1
○ Detailed results are shown for each Site I-IV, and for overall performance
○ Evaluations are done against other submissions, and benchmark methods, e. g. PIRADs v2.1 and IMPROD bpMRI Likert
○ In cases of continuous values is submitted, a Youden's index is calculated to binarise the submission
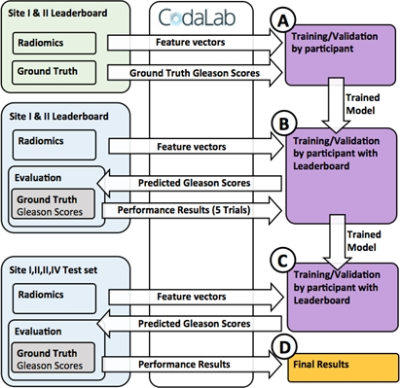
The challenge is implemented at freely available Codalab site (https://competitions.codalab.org), allowing download of training data, and running performance evaluations for submissions at different phases of the challenge (Figure 2). The performance evaluations are done with Area Under Receiver Operating Characteristic Curve (AUC), Sensitivity, Specificity, F1-score and Matthews Correlation Coefficient. The submissions are ranked according to AUC of patient-level classifications. The included radiomics contain exhaustive collection of all major publicly available techniques published for prostate cancer MR analysis with texture features in 2D and 3D, shape features, statistical descriptors, and topology metrics evaluted for repeatability [8], and pyradiomics radiomic feature extraction package [9]. For reference, programming code evaluating submission performance is given publicly available.
RESULTS & DISCUSSION
The challenge is available for use at https://competitions.codalab.org, and allows free evaluation of radiomic feature in with-in-site setting (Site I and II), and for two unseen sites (Site III and IV), simulating situation where a ML tool is developed using data from centers with which will use the ML tool as well centers which will not provide data for the development. This challenge is a part of series of public challenges consisting of 5 individual challenges:Challenge 1: ASCII files - Aim: Patient level class of GGG (0,1,2,3,4,5)
Challenge 2: Nifty files of T2w with whole gland masks - Aim: Segmentation of prostate on T2w.
Challenge 3: Nifty files of DWI (DWI5b500) with whole gland masks - Aim: Segmentation of prostate on DWI.
Challenge 4: Nifty files of T2w and DWI (DWI5b500) with whole gland masks and lesion masks - Aim: Patient level prediction of GGG (0,1,2,3,4,5) class.
Challenge 5: Nifty files of T2w and DWI (DWI5b500) with whole gland masks and lesion masks - Aim: A. Patient level prediction PIRADs version 2.1 (0,1,2,3,4,5) class; B. Lesion segmentation compared with radiologist’s lesion masks.
CONCLUSION
PRORAD challenge set is designed to provide independent evaluation of machine learning methods in dataset having uniform acquisition protocol in multiple sites.Acknowledgements
Organizers and acknowledgements
Organizers
Harri Merisaari, Department of Diagnostic Radiology, University of Turku and Turku University, Turku, Finland
Ivan Jambor, Department of Diagnostic Radiology, University of Turku and Turku University Hospital, Turku, Finland
Acknowledgements
Initials: H.J.A. Hannu Juhani Aronen; P.J.B. Peter J. Boström; O.E. Otto Ettala; I.J. Ivan Jambor; A.S. Aida Stainer; E.K. Esa Kähkönen; H.M. Harri Merisaari; I.M. Ileana Montoya Perez; O.S. Outi Oksanen; A.K. Antti Rannikko; J.R. Jarno Riikonen; M.S. Marjo Seppänen; K.S. Kari Syvänen; P.T. Pekka Taimen; P.V. Paula Vainio; J.V. Janne Verho; S. M. V. Sanna Mari Vimpeli
H.M., I.J.: Pre-processed the data and established the challenge.
H.M.: Developed and calculated the radiomic features.
P.J.B., O.E., E.K., A.R., J.R, K.S., M.S.: Responsible for patient recruitment.
I.J.: Prospectively reported and annotated all MRI data - individually T2-weighted imaging (T2w_tra) and Diffusion Weighted Imaging (DWI5b500, DWI2b1500, DWI2b2000) data sets.
H.M., I.J.: Quality Controlled the data.
P.J.B., O.E., E.K., A.R., J.R, K.S., M.S.: Performed biopsy procedures.
P.T., P.V.: Analyzed all histopathological (biopsy, prostatectomy) material.
P.J.B., O.E., I.J., E.K., H.M., O.O., A.R., J.R., M.S., K.S., J.V., S.M. V.: Took part in data collection.
I.J., I.M.: Constructed and maintain the online databases.
O.E., I.J., A.K.: Obtained ethical approval and registered the trials.
P.J.B., H.J.A., I.J.: designed the clinical trials.
P.J.B., H.J.A., I.J.: obtained funding for the trials.
This study was financially supported by grants from Instrumentarium Research Foundation, Sigrid Jusélius Foundation, Turku University Hospital, TYKS-SAPA research fund, Finnish Cultural Foundation, and Orion Research Foundation. HM was supported by the Cultural Foundation of Finland, and OrionPharma Research Fellowship.References
[1] Pesapane, F., Codari, M. and Sardanelli, F., 2018. Artificial intelligence in medical imaging: threat or opportunity: Radiologists again at the forefront of innovation in medicine. European radiology experimental, 2(1), p.35.
[2] Parmar, C., Barry, J.D., Hosny, A., Quackenbush, J. and Aerts, H.J., 2018. Data analysis strategies in medical imaging. Clinical cancer research, 24(15), pp.3492-3499.
[3] Sollini, M., Antunovic, L., Chiti, A. and Kirienko, M., 2019. Towards clinical application of image mining: a systematic review on artificial intelligence and radiomics. European journal of nuclear medicine and molecular imaging, pp.1-17.
[4] Prevedello, L.M., Halabi, S.S., Shih, G., Wu, C.C., Kohli, M.D., Chokshi, F.H., Erickson, B.J., Kalpathy-Cramer, J., Andriole, K.P. and Flanders, A.E., 2019. Challenges related to artificial intelligence research in medical imaging and the importance of image analysis competitions. Radiology: Artificial Intelligence, 1(1), p.e180031.
[5] Maier-Hein, L., Eisenmann, M., Reinke, A., Onogur, S., Stankovic, M., Scholz, P., Arbel, T., Bogunovic, H., Bradley, A.P., Carass, A. and Feldmann, C., 2018. Why rankings of biomedical image analysis competitions should be interpreted with care. Nature communications, 9(1), pp.1-13.
[6] Hoffmann, F., Bertram, T., Mikut, R., Reischl, M. and Nelles, O., 2019. Benchmarking in classification and regression. Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery, 9(5), p.e1318.
[7] Epstein, J.I., Zelefsky, M.J., Sjoberg, D.D., Nelson, J.B., Egevad, L., Magi-Galluzzi, C., Vickers, A.J., Parwani, A.V., Reuter, V.E., Fine, S.W. and Eastham, J.A., 2016. A contemporary prostate cancer grading system: a validated alternative to the Gleason score. European urology, 69(3), pp.428-435.
[8] Merisaari, H., Taimen, P., Shiradkar, R., Ettala, O., Pesola, M., Saunavaara, J., Boström, P.J., Madabhushi, A., Aronen, H.J. and Jambor, I., 2020. Repeatability of radiomics and machine learning for DWI: Short‐term repeatability study of 112 patients with prostate cancer. Magnetic Resonance in Medicine, 83(6), pp.2293-2309.
[9] Griethuysen, J. J. M., Fedorov, A., Parmar, C., Hosny, A., Aucoin, N., Narayan, V., Beets-Tan, R. G. H., Fillon-Robin, J. C., Pieper, S., Aerts, H. J. W. L. (2017). Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Research, 77(21), e104–e107.
Figures