4061
Deep Learning Reconstruction of 3D Zero Echo Time Magnetic Resonance Images for the Creation of 3D Printed Anatomic Models1Department of Radiology, Montefiore Medical Center, Bronx, NY, United States, 2Center for Advanced Imaging Innovation and Research, Department of Radiology, NYU Langone Health, New York, NY, United States, 3GE Healthcare, Waukesha, WI, United States, 4GE Healthcare, Munich, Germany, 5GE Healthcare, San Diego, CA, United States, 6GE Healthcare, Aurora, OH, United States, 7GE Healthcare, New York, NY, United States
Synopsis
Patient-specific three-dimensional (3D) printed anatomic models are valuable clinical tools which are generally created from computed tomography (CT). However, magnetic resonance imaging (MRI) is an attractive alternative, since it offers exquisite soft-tissue characterization and flexible image contrast mechanisms while avoiding the use of ionizing radiation. The purpose of this study was to evaluate the image quality and assess the feasibility of creating 3D printed models using a 3D Zero echo time (ZTE) MR images which were reconstructed with a deep learning reconstruction method.
Background
Three-dimensional (3D) printed models are valuable tools that may be used for many applications including pre-operative planning and simulations, real-time intra-operative surgical guides, and trainee and patient education.1 Although any type of volumetric imaging data may be used to create patient-specific 3D printed models, computed tomography (CT) is most commonly used due to the ease of performing image segmentation on CT data.2 Magnetic resonance imaging (MRI) is an attractive alternative, since it offers exquisite soft-tissue characterization and flexible image contrast mechanisms while avoiding the use of ionizing radiation. Segmentation of MRI data is challenging, however, due to noise, artifacts, magnetic field inhomogeneities, lower spatial resolution as compared to CT, and limitations of automated segmentation tools. The purpose of this study was to evaluate the image quality and assess the feasibility of creating 3D printed models using a 3D Zero echo time (ZTE) MR images which were reconstructed with a deep learning reconstruction method.Methods
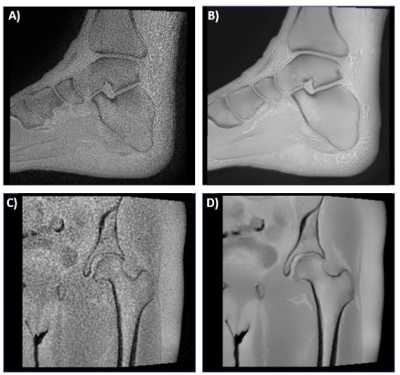
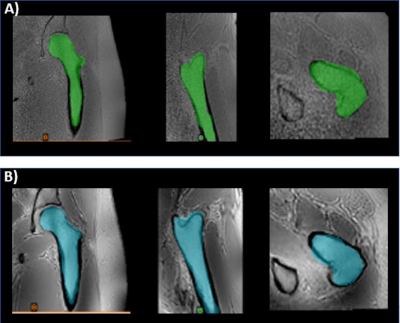
MRI of ten musculoskeletal (MSK) datasets was performed at 1.5T (Artist, GE Healthcare, Waukesha, WI) or 3T (Premier or Architect, GE Healthcare, Waukesha, WI) with and without deep learning. The datasets included two hips, three shoulders, two ankles, two c-spines, and one long bone (femur). Representative images from two of the datasets are shown in Figure 1. The 3D Zero TE (ZTE) method included a 3D radial acquisition to acquire the FID signal immediately after the RF excitation pulse. In this acquisition, the echo time is close to 0, enabling efficient capture of short T2 bone signals and provides flat PD contrast in soft-tissue in the background.3 A deep learning reconstruction method was also employed to improve signal-to-noise ratio of this acquisition. The Deep Learning Reconstruction method4 uses a deep convolutional residual encoder network trained to reconstruct images with reduction in noise. Signal intensity was inverted to provide CT like contrast. All results were blinded and randomized for image quality assessment by two radiologists both with over 30 years of experience and one radiology resident using a five-point scale (1=non-diagnostic, 2=poor, 3=adequate, 4=good, 5=excellent). A paired t-test was performed to compare mean scores (Matlab, Mathworks, Inc, Framingham, MA). DICOM images were imported to a dedicated 3D image-post processing platform (Mimics, Materialise, Leuven, Belgium) for 3D segmentation. The bones were segmented using a combination of automated and manual techniques (Figure 2), and the resultant 3D models were compared with and without deep learning.Results
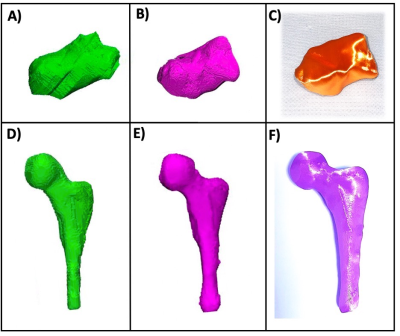
Deep learning significantly improved the image quality. The images with the deep learning reconstruction were less noisy and had enhanced edges between the bones and surrounding tissue. Mean scores were 2.73 ± 1.01 and 4.03 ± 1.03 for the non-deep learning and deep learning datasets respectively (p < 0.01). Automated segmentation tools such as level tracing, region growing, and edge detection worked better on the deep learning datasets as compared to the datasets with no deep learning which greatly facilitated the image segmentation. In addition, deep learning significantly improved the resultant 3D models created from the segmented regions of interest (Figure 3).Conclusions
This novel deep learning reconstruction method of MRI data provided excellent image quality compared to conventional MRI sequences and made it an excellent choice for the creation of 3D printed bone models. Having the ability to easily and accurately delineate bone from MRI data is projected to be especially useful for orthopedic oncology cases where MRI is necessary to visualize soft tissue structures; and 3D printed models including the bone, dominant lesion, and surrounding neurovasculature can help to identify the optimal surgical plan. Furthermore, performing 3D ZTE MRI with deep learning could potentially reduce the need for an additional CT exam which would lower costs and would save the patient from receiving extra ionizing radiation.Acknowledgements
No acknowledgement found.References
1] Mitsouras D, Liacouras P, Imanzadeh A, Giannopoulos A, Cai T, Kumamaru KK, George E, Wake N, Caterson EJ, Pomahac B, Ho V, Grant GT, Rybicki FJ. Medical 3D Printing for the Radiologist. RadioGraphics 2015, Nov-Dec; 35(7):1965-88.
[2] Rengier F, Mehndiratta A, von Tengg-Kobligk H, et al. 3D printing based on imaging data: review of medical applications. Int J Comput Assist Radiol Surg. 2010; 5(4):335-41.
[3] Weisinger F. et al. Zero TEMR bone imaging in the head. MRM 2016, vol 75 https://doi.org/10.1002/mrm.26094
[4] Lebel RM (2020) Performance characterization of a novel deep learning-based MR image reconstruction pipeline; arXiv:2008.06559
Figures