4057
Deep Learning Improves Detection of Anterior Cruciate Ligament- and Meniscus Tear Detection in Knee MRI1Department of Diagnostic and Interventional Radiology, Aachen University Hospital, Aachen, Germany, 2Institute of Imaging and Computer Vision, RWTH Aachen University, Aachen, Germany, 3Praxis im Köln Triangle, Cologne, Germany, 4Department of Diagnostic and Interventional Radiology, Düsseldorf University Hospital, Dusseldorf, Germany
Synopsis
In this study we aimed to analyze the capability of neural networks to accurately diagnose the presence of ACL and meniscus tears in our in-house dataset comprised of 3887 manually annotated knee MRI exams. To this end we trained the MRNet architecture on a varying number of training exams that included proton density-weighted axial, sagittal and coronal planes for each knee exam. Additionally, we compared the performance of the architecture when trained on expert vs non-expert annotations. This study demonstrates that while our neural network benefits from a larger dataset, expert annotations do not considerably improve the performance.
Purpose:
Anterior cruciate ligament- and meniscus tears are two very common injuries among athletes who participate in contact sports. Accurately diagnosing these injuries is of utmost importance for substantiated and timely therapeutic decision-making. Using magnetic resonance imaging (MRI), these injuries can be routinely diagnosed by experienced radiologists. Machine learning methods may help in detecting these injuries. Recent advances in computer vision, especially in the field of deep learning, have led to a myriad of new techniques that show promising results in recognizing various aspects of images. Especially in the realm of medical image analysis, several deep learning techniques have been successfully applied to accurately diagnose various injuries in common radiology exams such MRI, CT and X-Ray scans.1-3 In this study we want to build up on these improvements in order to evaluate the capability of neural networks to accurately detect ACL- and meniscus tears in our in-house dataset comprised of 3887 manually annotated knee MR exams.Methods
Our dataset comprises 3887 knee MR exams that were manually annotated with respect to the occurrence of ACL and meniscus tears. The labels used as the ground truth for each exam were set by medical students (non-experts), based on the previously written medical report by experienced radiologists. In addition, 246 of the 3887 knee MRI exams were also labeled by a board-certified radiologist (expert), in order to assess the performance difference between neural networks trained with labels generated by experts vs. non-experts. The overall dataset contains 1319 meniscus tears, 267 ACL tears as well as 2301 exams with no such injury and was split into a training set (2493 exams), a validation set (614 exams) and a test set (780 exams). Each exam consists of three knee MRI scans that cover the axial, coronal and sagittal planes using a fat-saturated proton density-weighted sequence.To allow our neural network to use all the available information present in the three planes, we based our architecture on the model proposed by Bien et al.4 that uses a separate neural network for each plane and injury and subsequently combines the results using logistic regression. We thus trained a total of nine neural networks that in a first step extracted the features of each input slice using the AlexNet5 architecture (pretrained on ImageNet6) followed by a global average pooling layer. The results of each slice were then combined by using a max pooling step across all slices. In a last step, a fully connected layer was used to predict the presence of either ACL or meniscus tears. To analyze the performance of the trained classifiers we employed the area under curve (AUC) of the receiver operating characteristic (ROC) curve as our metric of choice.
Results
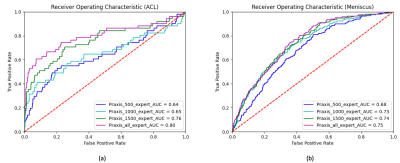
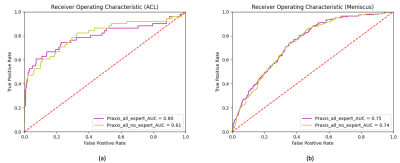
In a first experiment we used several training configurations, employing a different number of training samples to analyze the performance benefit of including more data in our training routine. Figure 1 shows the ROC curves for the cases where we trained neural networks with n=500, n=1000, n=1500 and all training samples (n=2493). For detecting ACL tears, the AUC constantly increased from 0.64 (n=500) to 0.80 (n=2493). For detecting meniscus tears, the AUC increased from 0.68 (n=500) to 0.75 (n=2493). In a second experiment we trained our neural networks with all available training data to study the influence of using expert annotations instead of student annotations in our training. Therefore, we trained a first network using only the expert labels in all cases that had been annotated by both an expert and a non-expert. Subsequently, we trained a second network that relied on labels set by the non-experts. The resulting ROC curves can be seen in Figure 2 (a) for ACL tears and Figure 2 (b) for meniscus tears. We found that both for ACL tear detection as well as for meniscus tear detection the additional expert annotations did not influence the network performance considerably.Discussion
Our results show that detection of ACL and meniscus tears is feasible using neural networks. We demonstrated that the AUC increases in step with the availability of larger training data. This suggests that in addition to looking for neural network architectures that utilize our dataset more efficiently, a sufficiently large dataset is mandatory to achieve human-level performance in clinical tasks. Moreover, we found that expert annotations for ACL and meniscus tears are not necessary in order to increase the performance of our classifier. This suggests that the detection of ACL and meniscus tears seem to be an easy problem for sufficiently trained non-experts and therefore allow the creation of a large dataset without the need for board-certified radiologists.Acknowledgements
No acknowledgement found.References
[1] Mohsen, Heba, et al. "Classification using Deep Learning Neural Networks for Brain Tumors." Future Computing and Informatics Journal 3.1 (2018): 68-71.
[2] Lakshmanaprabu, S. K., et al. "Optimal Deep Learning Model for Classification of Lung Cancer on CT Images." Future Generation Computer Systems 92 (2019): 374-382.
[3] Stephen, Okeke, et al. "An Efficient Deep Learning Approach to Pneumonia Classification in Healthcare." Journal of Healthcare Engineering 2019 (2019).
[4] Bien, Nicholas, et al. "Deep-Learning-Assisted Aiagnosis for Knee Magnetic Resonance Imaging: Development and Retrospective Validation of MRNet." PLoS Medicine 15.11 (2018): e1002699.
[5] Krizhevsky, Alex, Ilya Sutskever, and Geoffrey E. Hinton. "ImageNet Classification with Deep Convolutional Neural Networks." Communications of the ACM 60.6 (2017): 84-90.
[6] Deng, Jia, et al. "ImageNet: A large-scale Hierarchical Image Database." 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE, 2009.
Figures