4056
Self-Supervised Deep Learning for Knee MRI Segmentation using Limited Labeled Training Datasets1Stanford University, Stanford, CA, United States
Synopsis
Deep learning (DL)-based approaches have shown promise for automating medical image segmentation with high efficacy. However, current state-of-the-art DL supervised methods require large extents of labeled training images, which are difficult to curate at scale. In this work, we propose a self-supervised training scheme to reduce dependence on labeled data by pretraining networks in an unsupervised manner. We show that our method can improve segmentation performance, especially in the context of very limited data scenarios (only 10-25% scans available) and can achieve or surpass the accuracy of state-of-the-art supervised networks with approximately 50% fewer labeled scans.
Introduction
Segmentation of tissues from knee MRI scans have been used previously for evaluating clinical disorders and disease progression in osteoarthritis. The current gold-standard for segmentation uses manual methods that rely on expert readers. While accurate, these methods are cumbersome, time-consuming, and introduce intra- and inter-reader variations. While recent deep learning segmentation approaches have provided an excellent mechanism to automate this process, these techniques still rely on large annotated training datasets that can be problematic to curate.To overcome this challenge of requiring large labeled datasets for knee MRI segmentation, we explore self-supervised learning (SSL). SSL utilizes unlabeled data by first pretraining on a pretext task that has no downstream importance and does not require manual labels. This network can subsequently be tuned for the segmentation task of interest using limited labeled datasets. In this work, we analyze two pretext tasks, context prediction1 and context restoration2, to explore data-limited SSL segmentation to segment soft tissues from knee MRI scans.
Methods
155 quantitative double-echo steady-state (qDESS)3 knee MRI volumes from subjects referred for a diagnostic MRI scan were used in this work (sequence parameters described in the cited paper). Each scan consisted of 160 slices each and had expert manual segmentations for the femoral, tibial, and patellar cartilage, and the meniscus. The 155 scans were split into 86/33/36 scans for training/validation/testing.For SSL and simulating scans without any labels, the training set was split into 4 additional subsets to represent data-limited scenarios. The subsets consisted of a random 50%, 25%, 10%, and 5% of the training data (43, 22, 10, and 5 scans, respectively). All the training data, without labels, was used for pretext task training.
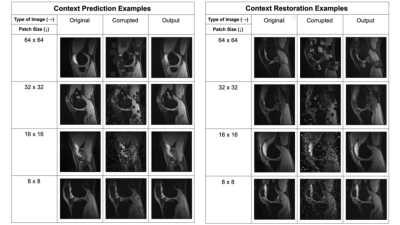
The two pretext tasks investigated in this work were context prediction and context restoration. For both tasks, the input 2D slice was corrupted and the network learned to fix this corruption through inpainting. In context prediction, randomly-selected patches in the input image are filled with zeros, while in context restoration, randomly-selected non-overlapping pairs of patches in the input image are swapped. We investigated four different patch sizes: 64-x-64, 32-x-32, 16-x-16, and 8-x-8 for both tasks. For each input image, patches were selected until the combined area of all selected patches exceeds 25% of the total image area. Example corruptions for both pretext tasks are shown in Figure 1.
The SSL pretext inpainting networks and the segmentation network utilized U-Nets4 with L2 loss and Dice loss, respectively. Each U-Net consisted of four levels with the number of channels increasing by a factor of 2 each level, from 32 to 256. Due to a small batch size, group normalization5 was used rather than batch normalization, and every convolutional layer directly preceding each group normalization layer included weight standardization6.
After training on the pretext tasks, we fine-tuned with labeled data with two training runs. First, all weights were frozen except the final convolutional layer for 100 epochs. Subsequently, all weights were unfrozen and the network was trained end-to-end for segmentation. Training was performed with an Adam optimizer, early stopping7, a batch size of 3, and with a learning rate of 1e-3 that decayed by 0.9 every 2 epochs.
We compared the segmentation performance of each combination of the two pretext tasks and four patch sizes, for the four data-limited scenarios. We also compared performance baselines with purely supervised training with the limited datasets and also to supervised training with all available labeled data.
Results and Discussion
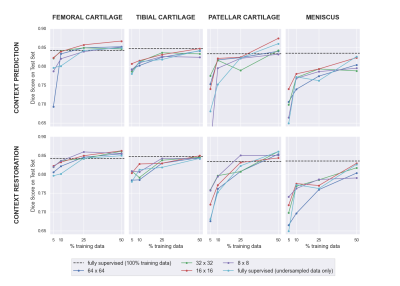
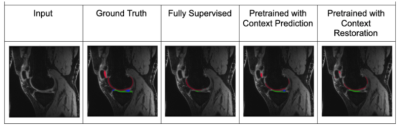
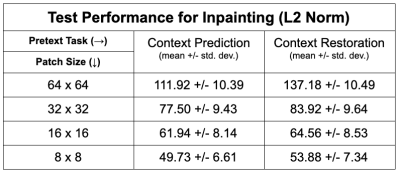
Of the two pretext tasks, inpainting for context prediction had better pre-training performance than context restoration, along with using smaller patch sizes for both (Table 1).In a data-limited regime, the use of SSL enabled using approximately only 10-25% labeled datasets without considerably degrading the performance of tissue segmentation, compared to 100% of the training data (Fig. 2). Most SSL networks exceeded the performance of only training with a limited extent of fully-supervised training data. Test performance varied more with respect to patch size when segmenting the patellar cartilage and the meniscus. Similarly, the improvement afforded by SSL was also higher in these sparser classes, which may suggest that SSL may be more beneficial for sparser classes of segmented tissues that suffer from class-imbalance.
A patch size of 16-x-16 performed the best or close to the best for all segmentation classes and both pretext tasks. Moreover, there were cases where multiple patch sizes work similarly well. Based on these observations, future work could explore ensembles of varying different patch sizes in order to generate more robust representations of the input images.
In some scenarios, fully supervised segmentation using 100% of the training data performed worse when segmenting the femoral and patellar cartilage than fully supervised segmentation using 50% of the data. These results suggest the need for training models with varying multiple random seeds to ensure robustness during deployment.
Conclusion
In this work, we explored the choice of pretext tasks and their hyperparameters for medical imaging. We demonstrate that the use of self-supervised learning to leverage unlabeled images can improve the segmentation performance of networks in scenarios with limited training data.Acknowledgements
We would like to acknowledge our funding sources: National Institutes of Health (NIH) grant numbers, NIH R01-AR077604, R00 EB022634, R01 EB002524, R01-AR074492, K24 AR062068, and P41 EB015891. GE Healthcare, Philips, and Stanford Medicine Precision Health and Integrated Diagnostics.References
1. Pathak, Deepak, et al. "Context encoders: Feature learning by inpainting." Proceedings of the IEEE conference on computer vision and pattern recognition. 2016.
2. Chen, Liang, et al. "Self-supervised learning for medical image analysis using image context restoration." Medical image analysis 58 (2019): 101539.
3. Chaudhari, Akshay S., et al. "Combined 5‐minute double‐echo in steady‐state with separated echoes and 2‐minute proton‐density‐weighted 2D FSE sequence for comprehensive whole‐joint knee MRI assessment." Journal of Magnetic Resonance Imaging 49.7 (2019): e183-e194.
4. Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
5. Wu, Yuxin, and Kaiming He. "Group normalization." Proceedings of the European conference on computer vision (ECCV). 2018.
6. Qiao, Siyuan, et al. "Weight standardization." arXiv preprint arXiv:1903.10520 (2019).
7. Prechelt, Lutz. "Early stopping-but when?." Neural Networks: Tricks of the trade. Springer, Berlin, Heidelberg, 1998. 55-69.
Figures