4046
Development of Deep Learning based Cartilage Segmentation at 3D knee MRI for the use of Biomarker of Osteoarthritis1Radiology, Korea University Guro Hospital, KUGH-MIDC, Seoul, Korea, Republic of, 2Korea University Anam Hospital, Seoul, Korea, Republic of, 3Korea University Ansan Hospital, Ansan, Korea, Republic of
Synopsis
Cartilage loss is fundamental pathology of knee osteoarthritis (OA). Quantitative analysis of cartilage thickness and volume is very time consuming by manual measurement. We proposed development of deep learning based cartilage segmentation at three dimensional knee magnetic resonance images, which can measure thickness and volume of knee joint cartilage, automatically and accurately. To evaluate the performance, we used Dice Similarity Coefficient (DSC) respect to the manual segmentation, and visual inspection. The accuracy DSC values were higher than 0.9. We expect deep learning program can be useful in future study for knee joint osteoarthritis.
Purpose
To develop and evaluate automated knee joint cartilage segmentation method using deep-learning technique in three dimensional magnetic resonance (MR) images, which can provide volume and thickness values of cartilage accurately, and rapidly.Method
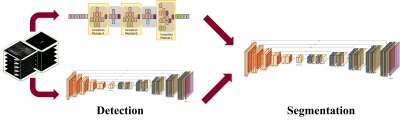
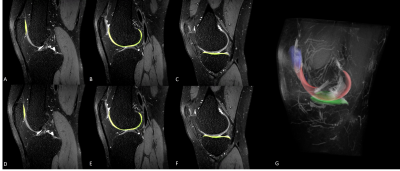
MRI data sets were obtained from 100 patients with Kellgren Lawrence grade 1-2, without previous knee joint surgical history, (68 men and 32 women, with an average age of 29.7 years and an age range of 12-71 years) who underwent a clinical MRI examination of the knee at our institution using the same 3.0-T MRI unit (Magnetom Skyra, Siemens Medical Solutions, Erlangen, Germany). The MRI data sets consisted of sagittal fat-suppressed proton density (PD) 3D CAIPIRINHA SPACE TSE sequence. The MRI performed with the following parameters: repetition time msec/echo time msec, 1000/45; field of view, 16 cm; matrix, 320 x 320; bandwidth, 390 Hz/pixel; and final image resolution, 0.56 x 0.56 x 0.5 mm. The segmentation method was developed based on deep-learning techniques with combining sagittal area detection model and segmentation model. The process was splited into two ways to solve the weight-imbalance problem and improve efficiency of the model. In detection phase, Inception V3 and UNET was used to determine the presence of knee joint cartilage. The modified U-net architecture based deep learning model with additional fully-connected layer was used for segmentation model. Multichannel images of the patient combining the 2.5-dimensional sagittal information were analyzed and used for the training dataset. The knee cartilage area was manually segmented by two trained radiology technicians under supervising of 2 musculoskeletal radiologists, using In-house developed software. The cartilage of patella, femur, and tibia were segmented separately. The volumetric image features of the segmented result were measured, such as thickness, and volume. The model was trained on 80, tested on 20 datasets. We used Dice Similarity Coefficient (DSC) to measure the performances of the automated methods, with respect to the manual segmentations.Result
The proposed segmentation method provided good performance for segmenting knee joint cartilage at each part of knee joint. The average accuracy/loss DSC values for patella was 0.962 / 0.174, for femur was 0.954 / 0.174, and for tibia was 0.937 / 0.177. The models average took less than 10 seconds to generate automatic segmentation in one dataset, using conventional personal computer with graphics processing units. In manual segmentation, it took about 4 hours to mask all cartilages in one dataset.Discussion
Our study described a fully automated cartilage segmentation system utilizing modified inception model and UNET for detecting knee joint cartilage, followed by modified UNET for segmenting cartilage tissue at each part of knee joint. All results for model evaluation (Dice coefficients, speed) are competitive with or outperform manual segmentation. Our study suggests the feasibility of using a deep leaning approach for fully automated model measuring the thickness and volume of articular cartilage of the knee joint, which can be quantitative biomarkers in future study evaluating osteoarthritis. While our deep learning model show good performance, there are several limitations. First, the number of our dataset is not enough to eliminate overfitting problem. Second, our accuracies are calculated assuming manual segmentation as the reference standard, inter-user variability must be concerned. Third, only three cartilage regions, femoral, tibia, and patella, were used in current method, whereas more detailed subregions of the cartilage can be inferred about osteoarthritis. In the next step, however, we could continue and develop this deep learning model for local area cartilage segmentation and volume, thickness measurement in the region of interest (ROI).Conclusion
The study demonstrates that U-net based deep-learning method is useful for rapid and accurate cartilage segmentation within knee joint, providing automated cartilage volume and thickness measurement of entire region in MR images, which can be used as a quantitative and objective biomarkers for osteoarthritis evaluation in future study.Acknowledgements
-References
1. Fang Liu, Zhaoye Zhou, Hyungseok Jang, et al. Deep Convolutional Neural Network and 3D Deformable Approach for Tissue Segmentation in Musculoskeletal Magnetic Resonance Imaging. Magn Reson Med. 2018 Apr;79(4):2379-2391
2. F. Eckstein, J.E. Collins, M. C. Nevitt, et al. Cartilage Thickness Change as an Imaging Biomarker of Knee Osteoarthritis Progression: Data From the Foundation for the National Institutes of Health Osteoarthritis Biomarkers Consortium. Arthritis & Rheumatology 2015 Dec;67(12):3184-9.
3. L F Schaefer, M Sury, M Yin, et al. Quantitative measurement of medial femoral knee cartilage volume e analysis of the OA Biomarkers Consortium FNIH Study cohort. Osteoarthritis and Cartilage 2017 Jul;25(7):1107-1113
Figures