4001
Whole brain CSF segmentation for consistent zero-referencing and longitudinal study applicability of MEDI+01Radiology, Weill Cornell Medicine, New York, NY, United States, 2Neurology, Weill Cornell Medicine, New York, NY, United States, 3Weill Cornell Medicine, New York, NY, United States
Synopsis
The role of CSF segmentation for zero-referenced MEDI+0 is demonstrated. In the present study we show that it is essential to enforce consistency in the CSF mask between different timepoints to ensure value reproducibility and to minimize effects of random shifts in the solution of MEDI+0 minimization
Introduction
Quantitative susceptibility mapping (QSM) allows quantitative MRI-based estimation of distribution of magnetic susceptibility sources (calcium, iron, contrast agents) within a given tissue from complex gradient echo data. However, due to singularity of dipole kernel at the k-space center, reconstructed susceptibility maps are meaningful only with respect to a chosen reference. To make QSM applicable to absolute quantification of magnetic sources, CSF referencing has been proposed [1]. Standard implementation of MEDI+0 involves automated preparation of connectivity-filtered ventricular CSF mask – a method which might be susceptible to variations due to imaging resolution, sequence parameters, vendor, etc. These small differences may lead to differences in CSF segmentation, and, as a result, global shift in the solution of the MEDI+0 minimization, which limits the technique’s applicability in longitudinal studies. In here we investigate dependence of MEDI+0 on CSF mask structure, and propose whole brain CSF as a way to produce consistent susceptibility measurements in longitudinal data.Methods
Ten healthy subjects (3 male, 7 female, mean age 26.8 years ± 4.1) were imaged in two consecutive MRI sessions using the same acquisition protocol at 3T (Siemens Prisma VE11B scanner). There was a minimal delay between the scanning sessions, during which the subjects were asked to sit up and lie down again to mimic head repositioning in an actual MRI exam. The acquisition protocol included 1mm isotropic MPRAGE sequence for structural imaging and multi-echo GRE sequence for QSM (TE1/ΔTE = 6.3/4.06ms TR=48ms, FA=15°, voxel size = 0.8mm×0.8mm×3mm). R2* maps were calculated from the GRE magnitude images using ARLO [2] (taken from the MEDI Toolbox [3]) and thresholded at 5Hz to segment the $$$CSF_{brain}$$$ across the brain [1], resulting in a mask . Further image processing was performed (including connectivity analysis) to obtain the CSF within the ventricles or $$$CSF_{vent}$$$ using the implementation of MEDI+0 in the MEDI Toolbox. Sixteen subcortical ROIs (frontal WM, parietal WM, temporal WM occipital WM, thalamus, caudate nucleus, putamen and pallidum in both hemispheres) were automatically segmented using FreeSurfer [3] and coregistered onto GRE image using FMRIB’s FLIRT [4]. Brain susceptibility maps were reconstructed for each scan and each CSF reference mask. Repeatability of the of susceptibility values estimated using the two CSF reference masks were compared using Bland-Altman analysis. Brain susceptibility maps were reconstructed for each of the CSF reference masks in five MS patient (6-8 scans since 2011/2012). Lesions were manually traced by an expert reader on the baseline scan. Susceptibility time course of each lesion with respect to neighboring normal appearing white matter was recorded.Results
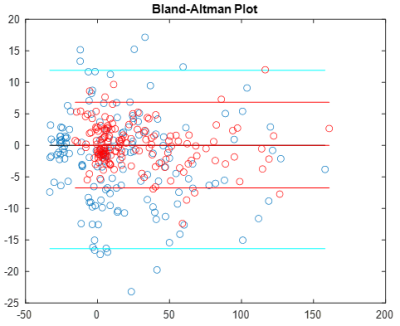
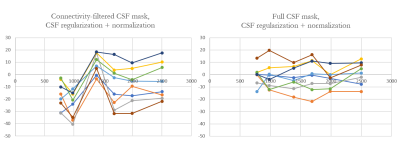
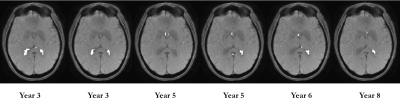
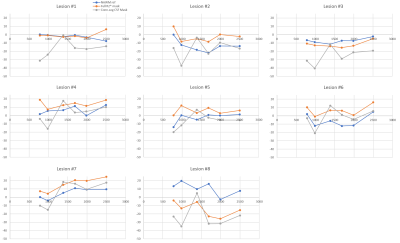
No significant volume differences for each segmented subcortical structure were detected each scan session. Furthermore, no significant differences in mean values of susceptibility were for each session for each reconstruction method. Fig. 1 demonstrates Bland-Altman plot of inter-scan agreement in measured ROI averages for standard ($$$CSF_{vent}$$$, blue) and full ($$$CSF_{brain}$$$, red) CSF masks across all ROIs and all subjects. While both methods demonstrate good agreement and lack of systematic bias, reconstruction utilizing was found to have significantly smaller limits of agreement. Figure 2 demonstrates time courses of 8 MS lesions obtained from one of the MS subjects estimated from the standard MEDI+0 reconstructions whit the mask $$$CSF_{vent}$$$. Noticeable synchronous jumps in susceptibility values can be observed. Inspection of the masks revealed large inconsistencies between the different timepoints (Fig 3). Fig 2b shows a comparison of lesion susceptibility time courses estimated from the QSM reconstructions referenced to the full $$$CSF_{brain}$$$ mask. Removal of the CSF mask’s connectivity filtering resulted in consistent reconstruction of the susceptibility values, reflective of the changes detected through local referencing. Fig 4 shows comparison of the lesion time courses estimated by different methods for each individual lesion.Discussion and Conclusion
Improved interscan agreement of QSM across multiple repeated scan sessions at a single 3T scanner was demonstrated for full CSF mask, suggesting potential utility in longitudinal studies and clinical applications. This consistency likely originates from the fact that larger regularization volume of the proposed full mask results in reduced sensitivity to variation in segmented volume.Acknowledgements
No acknowledgement found.References
[1.] Liu Z, Spincemaille P, Yao Y, Zhang Y, Wang Y. MEDI+0: Morphology enabled dipole inversion with automatic uniform cerebrospinal fluid zero reference for quantitative susceptibility mapping. Magn Reson Med. 2018;79(5):2795-2803.
[2.] Pei M, Nguyen TD, Thimmappa ND, et al. Algorithm for fast monoexponential fitting based on Auto-Regression on Linear Operations (ARLO) of data. Magn Reson Med. 2015;73(2):843-850.
[3.] http://pre.weill.cornell.edu/mri/pages/qsm.html
[4.] Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774-781.
[5.] Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17(2):825-841.
Figures