3982
Automatic, Non-Regularized Nonlinear Dipole Inversion for Fast and Robust Quantitative Susceptibility Mapping1Department of Medical Physics and Biomedical Engineering, University College London, London, United Kingdom
Synopsis
Quantitative Susceptibility Mapping (QSM) is an ill-posed inverse problem often solved by regularization: minimizing a functional until it converges. This is usually time-consuming, requiring fine-tuning of several parameters by many repetitions of the whole optimization solver. Nonlinear Dipole Inversion is a QSM method that solves a nonlinear Tikhonov-regularized functional with a gradient descent solver. We show that stopping this method early provides optimal results, largely independent of the regularization weight. Here, we propose a non-regularized nonlinear conjugate gradient solver with a new stopping criterion based on analysing susceptibility map spatial frequency coefficients to achieve fast, parameter-free and automatic QSM.
PURPOSE
Susceptibility maps can be estimated from the phase of Gradient Echo MRI acquisitions by calculating local magnetic field maps and then solving an ill-posed inverse problem. Bayesian approaches to solving the inverse problem rely on minimizing a functional until it converges1. This is time-consuming and requires fine-tuning of one or more parameters by solving the functional many times. Most functionals are composed of one or more regularization terms which are balanced with a data-fidelity term via a Lagrangian weight ($$$\lambda$$$). For example, the linear QSM functional for Tikhonov regularization is2: $$$argmin_{\chi}\frac{1}{2}||W(F^HDF\chi -\phi)||^2_2+\lambda||\chi||^2_2$$$, with W a magnitude or SNR-based whitening weight, $$$\phi$$$ the local phase, and $$$F$$$ and $$$F^H$$$ the direct and inverse Fourier transforms. D is the dipole kernel in the frequency domain ($$$D=\frac{1}{3}-\frac{k_z^2}{k^2}$$$)3,4, and $$$\chi$$$ the optimized susceptibility map.A recent nonlinear gradient descent algorithm (Nonlinear Dipole Inversion, NDI)5 gives results comparable to state-of-the-art methods such as MEDI6 and FANSI7 and largely independent of the regularization weight. By stopping the NDI algorithm early, before it diverges from the true solution, it is possible to avoid the use of a regularization term entirely5,8. In this paradigm, the number of iterations becomes the main parameter that controls the results. Unfortunately, the gradient descent algorithm used in NDI typically requires hundreds of iterations (albeit each taking a fraction of a second) to achieve optimal results. More problematically, manual supervision is currently needed to stop NDI early as there are no automatic methods available to reliably stop the gradient descent algorithm. Here, we propose an efficient nonlinear conjugate gradient method8 to achieve optimal results in much fewer iterations than the gradient descent method. Furthermore, we show that by analyzing the spatial frequency coefficients9 of the susceptibility map reconstructed at each iteration, a heuristic rule can be determined to stop the algorithm before noise is amplified, thus preventing streaking artifacts in the final susceptibility map.
METHODS
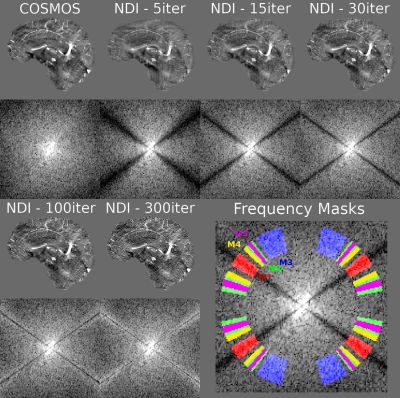
We aimed to solve the nonlinear Quantitative Susceptility Mapping (QSM) functional $$$argmin_{\chi}\frac{1}{2}||W(e^{iF^HDF\chi }-e^{i\phi})||^2_2$$$ with a nonlinear conjugate gradient solver. Exploiting the way that each iteration affects the solutions in the frequency domain9 (Figure 1), we analyzed this domain to determine the optimal number of iterations. We propose to use two new frequency masks ($$$M4: 0.15<|D|<0.20$$$ and $$$M5: 0.225<|D|<0.275$$$, with D the dipole kernel coefficients3,4) to stop the solver when the mean absolute value of the susceptibility map frequency coefficients inside M4 is larger than in M5.Two benchmark, synthetic brain datasets10,11 were used to compare the gradient descent (NDI, original formulation5) and nonlinear conjugate gradient (NDI_CG) algorithms ,both with automatic stopping, against state-of-the-art nonlinear total-variation (TV)-regularized FANSI7. Noisy local phase maps were simulated using the forward model3,4 from two numerical susceptibility phantoms: 1) A COSMOS reconstruction from the 2016 QSM challenge (RC1)10 (with SNR=100 and 300) and 2) The 2019 QSM challenge SIM2SNR111 dataset (RC2). For RC2, the FOV was doubled in every direction to prevent large-scale aliasing for all algorithms (final matrix size: 328x410x410). Reconstructions were compared in terms of normalized root mean squared error (RMSE), QSM-tuned structural similarity index (XSIM)12, computation time (i7-9750H @4.5GHz with 32Gb RAM), and the number of iterations. All reconstruction strategies were also tested on a challenging 3T in-vivo head and neck dataset13: 1.25 mm isotropic voxels, 4 echoes TE / dTE / TR = 4.61 ms / 4.61 ms / 22 ms , α = 12°, Tacq = 6 min 4s. Preprocessing included magnitude-thresholded masking, SEGUE14 unwrapping and MSHARP15 background field removal.
RESULTS
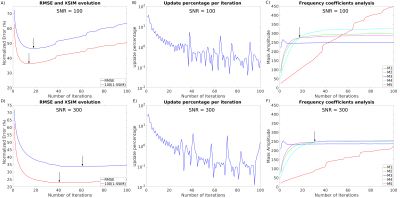
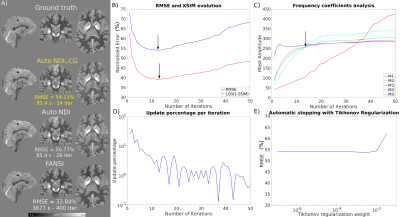
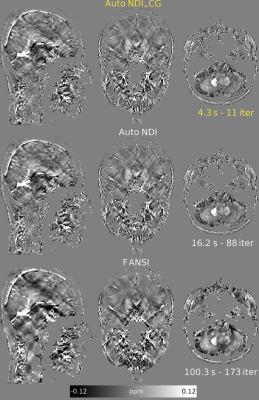
Figures 2 and 3 show the results using the RC1 simulations. At each noise level, the optimal number of iterations according to the minimum RMSE and maximum XSIM is very similar to the number of iterations at the intersection of the M4 and M5 amplitude curves where NDI_CG is stopped. Reconstructions using RC2 (Figure 4) also show the same agreement. The update percentage between iterations ($$$100(||\chi_k-\chi_{k-1}||)/||\chi_k||$$$ where $$$\chi_k$$$ is the susceptibility estimated at iteration k) was not a good indicator of the optimal number of iterations. Inserting a Tikhonov regularization term did not improve the RMSE for automatic NDI_CG (Figure 4E). Optimal reconstructions of the in-vivo dataset (Figure 5) show that automatic NDI_CG gives very similar susceptibility maps to NDI.DISCUSSION
The automatically stopped NDI_CG solver was much faster than traditional NDI, providing similar QSM RMSE/XSIM. Automatic NDI_CG is also faster than FANSI, particularly for large datasets. Both NDI methods gave worse RMSE than FANSI because no denoising is performed in NDI. Our automatic stopping criterion removes the need for time-consuming manual or L-curve-based optimization. In vivo, stopping the NDI_CG algorithm early gave no discernible susceptibility underestimation. This fast, automatic technique may be used in the future to accelerate regularized QSM by initialization or preconditioning16, or to find optimal regularization weights such as in bi-level approaches17.CONCLUSION
We have shown that it is possible to use a non-regularized nonlinear dipole inversion algorithm with an automatic stopping criterion, based on the analysis of susceptibility map frequency coefficients, to achieve a fast, parameter-free QSM reconstruction. In conjunction with a conjugate gradient solver, this provides a fast and robust estimation of susceptibility maps that reduces artifact propagation and noise amplification.Acknowledgements
Dr Carlos Milovic is supported by Cancer Research UK Multidisciplinary Award C53545/A24348. Dr Karin Shmueli is supported by European Research Council Consolidator Grant DiSCo MRI SFN 770939. We thank Dr. Anita Karsa for her MRI data and contributions.References
1. Kee Y, Liu Z, Zhou L, Dimov A, Cho J, de Rochefort L, Seo JK, Wang Y. Quantitative Susceptibility Mapping (QSM) Algorithms: Mathematical Rationale and Computational Implementations. IEEE Trans Biomed Eng. 2017;64:2531-2545;
2. Kressler B, Rochefort L De. Nonlinear regularization for per voxel estimation of magnetic susceptibility distributions from MRI field maps. IEEE Trans Med Imaging 2010;29:273–281;
3. Salomir R, De Senneville BD, Moonen CTW. A fast calculation method for magnetic field inhomogeneity due to an arbitrary distribution of bulk susceptibility. Concepts Magn Reson. 2003;19B:26-34
4. Marques J P, Bowtell R. Application of a Fourier-based method for rapid calculation of field inhomogeneity due to spatial variation of magnetic susceptibility. Concepts Magn Reson Part B Magn Reson Eng. 2005;25:65-78
5. Polak, D, Chatnuntawech, I, Yoon, J, et al. Nonlinear dipole inversion (NDI) enables robust quantitative susceptibility mapping (QSM). NMR Biomed. 2020;e4271
6. Liu T, Wisnieff C, Lou M, Chen W, Spincemaille P, Wang Y. Nonlinear formulation of the magnetic field to source relationship for robust quantitative susceptibility mapping. Magn Reson Med. 2013;69:467-476
7. Milovic C, Bilgic B, Zhao B, Acosta-Cabronero J, Tejos C. Fast nonlinear susceptibility inversion with variational regularization. Magn Reson Med. 2018;80:814-821
8. Milovic C, Karsa A, and Shmueli K. Efficient Early Stopping algorithm for Quantitative Susceptibility Mapping (QSM). ESMRMB 2020, Virtual Meeting.
9. Milovic C,Prieto C, Bilgic B, Uribe S, Acosta-Cabronero J, Irarrazaval P, Tejos T. Comparison of Parameter Optimization Methods for Quantitative Susceptibility Mapping. Magn Reson Med. 2020;85:480– 494
10. Langkammer C, Schweser F, Shmueli K, Kames C, Li X, Guo L, Milovic C, Kim J, Wei H, Bredies K, Buch S, Guo Y, Liu Z, Meineke J, Rauscher A, Marques JP, Bilgic B. Quantitative susceptibility mapping: Report from the 2016 reconstruction challenge. Magn Reson Med. 2018;79:1661-1673
11. Marques JP, Meineke J, Milovic C, Bilgic B, Chan K-S, Hedouin R, van der Zwaag W, Langkammer C, and Schweser F. QSM Reconstruction Challenge 2.0–Part 1: A Realistic in silico Head Phantom for MRI data simulation and evaluation of susceptibility mapping procedures. bioRxiv 2020.09.29.316836; doi: https://doi.org/10.1101/2020.09.29.316836
12. Milovic C, Tejos C, and Irarrazaval P. Structural Similarity Index Metric setup for QSM applications (XSIM). 5th International Workshop on MRI Phase Contrast & Quantitative Susceptibility Mapping, Seoul, Korea, 2019
13. Karsa A, Punwani S, and Shmueli K. An optimized and highly repeatable MRI acquisition and processing pipeline for quantitative susceptibility mapping in the head-and-neck region. Magn Reson Med. 2020; 84: 3206– 3222
14. Karsa A, Shmueli K. SEGUE: A Speedy rEgion-Growing Algorithm for Unwrapping Estimated Phase. IEEE Trans Med Imaging. 2019;38:1347-1357
15. Milovic C, Langkammer C, Uribe S, Irarrazaval P, Acosta-Cabronero J, and Tejos C. Multiscale Spherical Mean Value based background field removal method for Quantitative Susceptibility Mapping. 27th International Conference of the ISMRM, Montreal, Canada, 2019.
16. Liu Z, Kee Y, Zhou D, Wang Y, Spincemaille P. Preconditioned total field inversion (TFI) method for quantitative susceptibility mapping. Magn Reson Med. 2017;78:303-315
17. De los Reyes JC, Schönlieb CB. and Valkonen T. Bilevel Parameter Learning for Higher-Order Total Variation Regularisation Models. J Math Imaging Vis 2017;57:1–25
Figures