3887
Automatic Segmentation and Normal Dataset of Fetal Body from Magnetic Resonance Imaging
Bella Fadida-Specktor1, Dafna Ben Bashat2,3, Daphna Link Sourani2, Netanell Avisdris1,2, Elka Miller4, Liat Ben Sira3,5, and Leo Joskowicz1
1School of Computer Science and Engineering, The Hebrew University of Jerusalem, Haifa, Israel, 2Sagol Brain Institute, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 3Sackler Faculty of Medicine & Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel, 4Medical Imaging, Children’s Hospital of Eastern Ontario, University of Ottawa, Ottawa, ON, Canada, 5Division of Pediatric Radiology, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel
1School of Computer Science and Engineering, The Hebrew University of Jerusalem, Haifa, Israel, 2Sagol Brain Institute, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 3Sackler Faculty of Medicine & Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel, 4Medical Imaging, Children’s Hospital of Eastern Ontario, University of Ottawa, Ottawa, ON, Canada, 5Division of Pediatric Radiology, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel
Synopsis
Weight estimation is of great importance in assessing fetal development, yet unavailable in routine fetal MRI. The aim of this study was to develop an automatic fetal body segmentation method and to create a large dataset of volumetric body measurements of normal fetuses. Automatic fetal body segmentation was performed on data obtained from two clinical sites, three MRI systems and two sequences. Using a neural network trained for each sequence, high performance was achieved for both of them. A database of normal fetal volumes with a wide range of gestational age was created and was consistent with ultrasound growth chart.
Introduction
Fetal body weight estimation, routinely performed with ultrasound imaging, is of great importance for evaluating fetal development and pregnancy management. However, these estimations are based on manually acquired 2D linear measurements on ultrasound images, which have high user variability and usually overestimate the fetal body1. Fetal MRI, which is increasingly used for fetal development assessment, has the potential to yield an accurate and reliable fetal body volume measurements. However, this requires the delineation of the fetal body contour (segmentation), which is not performed in routine clinical practice since it is very time consuming (>1 hour per scan). Recently, both semi-automatic and automatic segmentation methods have been proposed2,5,6,7. However, these methods were limited to a specific sequence, and have not been applied to a large clinical cohort. This study proposes a fully automatic 3D segmentation method of the fetal body, developed for two MRI sequences, and generates for the first time volumetric normal fetal body measurements with a wide range of gestational age (GA).Methods
Subjects: MRI data for a total of 212 fetuses, with GA range 19-39 weeks, was included in this study. Of those, 101 fetuses had normal development and 111 had various pathologies.MRI Protocol: Data was obtained from two clinical sites with 3 MRI systems: 3T and 1.5T GE, and 3T Siemens, and included FIESTA and TRUFI sequences. FIESTA scans had in-plane resolution of 0.59-1.87mm and slice thickness of 3-7mm, and TRUFI scans had in-plane resolution of 0.59-1.34mm and slice thickness of 3-4mm.
Scans annotation: Initially, 64 scans were manually segmented. Subsequently, to facilitate creation of a large data cohort, 148 scans were obtained by correcting the network results. All annotations were approved by a senior pediatric-radiologist or expert neuro-scientist in fetal MRI.
Preprocessing: Images were normalized to zero mean and standard deviation of one. To increase the size of the dataset, extensive data augmentations were performed, including flipping, rotation, scaling, translation, elastic transformations, Poisson noise addition and contrast variations.
Network architecture and training: The segmentation method was developed on 187 annotated cases. A custom 3D U-Net network2 was used to train two fetal body segmentation networks, one for each sequence. For the FIESTA network, we used 45 cases for training, 6 for validation and 77 for testing. For the TRUFI network, we used 36 cases for training, 3 for validation and 20 for testing. Since most TRUFI scans had high resolution, we first downscaled them half in-plane and then trained TRUFI network on them as reported previously3. The training of FIESTA network was done without pre-training, while the TRUFI network was fine-tuned from FIESTA network because there were fewer scans in the training set. Both networks were trained with the Adam optimizer, initial learning rate of 0.0005 and batch size of 2. At test time, the scans were automatically rescaled in all dimensions to fit median resolution of the training set.
Implementation: Image analysis, network construction and training were performed using Python with Keras and Tensorflow frameworks.
Evaluation: Segmentation network results were evaluated using Dice score, the Hausdorff distance and the Asymmetric Surface Distance (ASSD) metrics.
Fetal total body volumetric database: A dataset of 101 volumetric measurements of normal fetuses was computed from the fetal body segmentations. A plot of fetal volumes with respect to fetal GA was then derived and compared to growth chart from ultrasound4.
Results
Networks Results: Networks performance is presented in Table 1. The FIESTA body segmentation network reached a Dice score of 0.977 on 77 test cases, indicating the robustness of our approach to variations in FIESTA parameters. TRUFI body segmentation network reached 0.975 Dice score on a test set of 20 cases, showing that our approach resulted in high quality segmentation for TRUFI protocol as well. Figure 1 illustrates representative segmentation results from FIESTA and TRUFI networks.Normal fetal body MRI database – comparison to ultrasound growth chart: Figure 3 shows normal fetal volumes as a function of GA plotted on top of the ultrasound growth chart4. As can be seen, most of the points lie within the ultrasound normal range, with some points slightly below the chart. The presence of a relatively large number of smaller fetuses may be due to our inaccurate assumption of specific gravity of 1g/cc8 and/or an over-estimation of the weight assessment using US1. Preliminary results of two fetuses diagnosed with fetal growth restriction (FGR) are shown in Figure 3. As expected, both lie significantly below the ultrasound growth chart and much below our MRI data points of similar GA.
Discussion & Conclusion
In this study we presented a fully automatic fetal body segmentation network that was shown to yield accurate fetal total body volumetric measurements on a large dataset of fetal MRI scans from TRUFI and FIESTA sequences. Network results greatly facilitated the annotation process. From a unified cohort of annotated data we further constructed for the first time a body growth dataset of normal fetuses in MRI. Our results are in line with ultrasound fetal growth chart and can be used as reference values for fetal assessment using MRI. In the future we are planning to enlarge our dataset and draw fetal growth chart.Acknowledgements
This work was supported by Kamin grants of the Israel Inovation Authority.References
- Milner, Julia, and Jane Arezina. "The accuracy of ultrasound estimation of fetal weight in comparison to birth weight: a systematic review." Ultrasound 26.1 (2018): 32-41.
- Dudovitch, Gal, et al. "Deep Learning Automatic Fetal Structures Segmentation in MRI Scans with Few Annotated Datasets." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2020.
- Isensee, Fabian, et al. "nnu-net: Self-adapting framework for u-net-based medical image segmentation." arXiv preprint arXiv:1809.10486 (2018).
- Kiserud, Torvid, et al. "The World Health Organization fetal growth charts: a multinational longitudinal study of ultrasound biometric measurements and estimated fetal weight." PLoS medicine 14.1 (2017): e1002220.
- Zhang, T., Matthew, J., Lohezic, M., Davidson, A., Rutherford, M., Rueckert, D., ... & Aljabar, P. (2016). Graph-based whole body segmentation in fetal MR images. In MICCAI workshop on PIPPI.
- Anquez, J., Bibin, L., Angelini, E. D., & Bloch, I. (2010, April). Segmentation of the fetal envelope on ante-natal MRI. In 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro (pp. 896-899). IEEE.
- Bibin, L., Anquez, J., Angelini, E., & Bloch, I. (2010). Hybrid 3D pregnant woman and fetus modeling from medical imaging for dosimetry studies. International journal of computer assisted radiology and surgery, 5(1), 49.
- Schild, R. L. (2007). Three‐dimensional volumetry and fetal weight measurement. Ultrasound in Obstetrics and Gynecology: The Official Journal of the International Society of Ultrasound in Obstetrics and Gynecology, 30(6), 799-803.
Figures
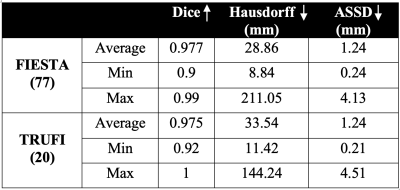
Table 1: Results of the fetal body segmentation networks for the FIESTA and TRUFI sequences.
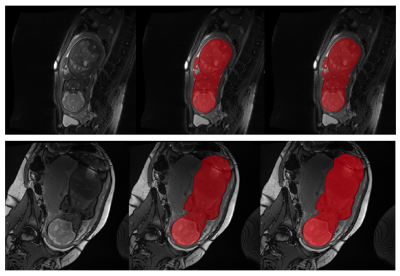
Fig. 1: Illustrative examples of the FIESTA (upper row) and TRUFI (lower row) fetal body segmentation results of two cases. Left: original image, middle: body segmentation result, right: manual ground truth.
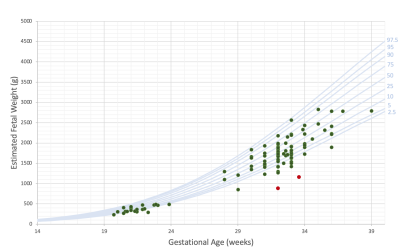
Fig. 2: Estimated fetal volume as a function of gestational age (green points) overlaid on ultrasound fetal body growth chart4 (blue curves representing percentiles). Red points correspond to IUGR fetuses.