3815
MRS metabolomics screening of human lung cancer with blood serum collected prior to disease diagnosis
L Cheng1, Tjada Schult1, Mara Lauer1, Yannick Berker2, Marcella Cardoso1, Lindsey Vandergrift1, Piet Habbel3, Johannes Nowak4, Martin Aryee1, Mari Mino-Kenudson1, and David Christiani5
1Mass General Hospital, Boston, MA, United States, 2German Cancer Research Center, Heidelberg, Germany, 3Charite Medical University, Berlin, Germany, 4Julius-Maximilians University, Wuerzburg, Germany, 5Harvard T.H. Chan School of Public Health, Boston, MA, United States
1Mass General Hospital, Boston, MA, United States, 2German Cancer Research Center, Heidelberg, Germany, 3Charite Medical University, Berlin, Germany, 4Julius-Maximilians University, Wuerzburg, Germany, 5Harvard T.H. Chan School of Public Health, Boston, MA, United States
Synopsis
Lung cancer (LuCa), the leading cause of cancer deaths, are often diagnosed late due to the lack of screening. Low-dose spiral CT can detect small and early stage LuCa lesions, but cannot practically be used as an annual LuCa screening tool. Metabolomics detects global metabolite variations under physiology and pathology. Metabolomic profiles measured from blood may reveal LuCa at early stages as a screening tool to triage suspicious patients to CT tests. Blood sera obtained from LuCa patients prior to their diagnosis were studied with MRS to establish LuCa screening metabolomic profiles to discover LuCa earlier and reduce death rates.
INTRODUCTION
Lung cancer (LuCa) is the leading cause of oncologic-related deaths. Early stages are mostly asymptomatic and contribute to a delayed diagnosis, with over 70% of the patients dying from LuCa. While the overall five-year survival rate is 19%, stage I LuCa can achieve a much higher five-year survival rate of 71%, which emphasizes the essential need for an effective screening test. At present, the best possible LuCa early detection test is a low-dose spiral CT, but due to its costs and radiation hazard it can hardly be used as a widespread screening method. Thus, a simple, preferably portable, non- or minimal-invasive screening test with no harmful side-effects is urgently needed to triage patients with suspicious screening results to a further CT test and thereby minimize LuCa associated mortalities.Cancer metabolomics detects oncological developments by interrogating the metabolic profiles from metabolic pathways through global metabolite variations. Previously published human serum MRS-based metabolomic profiles have shown abilities to differentiate LuCa from controls and between different cancer type. These successes encouraged us to study LuCa patient serum samples prior to their LuCa diagnosis to evaluate their capability as a screening agent.
METHODS
Samples. Sera from non-small cell LuCa (NSCLC)patients and their age, gender and smoking habit matched healthy controls were grouped according to the design of training-testing-validation cohorts in this study. The training cohort included 25 NSCLC sera from patients at the time of diagnosis (Time-of-Dx) and controls; the testing cohort consisted 25 sera collected 0.5 to 5 yrs Prior-to-Dx from the 25 NSCLC patients in the training cohort; and the validation cohort recruited sera collected less than 2 yrs Prior-to-Dx from additional 54 NSCLC patients and controls.MR Spectroscopy. High resolution magic angle spinning (HRMAS) MRS analysis of serum samples are performed at 4°C by a 600MHz Bruker spectrometer at 3,600Hz spinning rate. Spectra are analyzed with a MatLab-based curve fitting program.
Data Analysis. 57 spectral regions were selected based on the training and testing cohorts. Following selections of these regions, all of the data analytical procedures, including principal component and canonical analyses, were performed on the training cohort and followed by the testing and validation cohorts.
RESULTS
MRS of native sera (10ul) were measured with HRMAS MRS. Spectra presented as group averages with standard deviations for training and testing cohorts are shown in Fig. 1. Following selections of 57 spectral regions, PCA and canonical analysis were conducted on the training cohort with the aim to differentiate Time-of-Dx from Healthy groups, with the testing cohort passively followed the calculations. Results thus obtained from the training and testing cohorts are presented in Fig 2.Using the mean plus one standard error (M+SE) as the threshold, calculated from the canonical score differences between Time-of-Dx and Prior-to-Dx for each case, i.e. the difference of the two scores for each patient, a Kaplan-Meier survival analysis indicates significantly better survival rate from their time of NSCLC diagnoses for patients with score differences higher than the threshold (Fig 3a). Furthermore, within the group of stage I and IIA, patient survivals can be significantly predicted from the date of their Prior-to-Dx blood sample collections, if their score values are higher than the M+SE threshold, calculated by the testing cohort (dashed line in Fig 2), as shown in Fig 3b.
Fig 4a presents score results for cases in the validation cohort as comparisons with those in the training and testing cohorts. The validation cohort demonstrated the same significant trend of score changes in the NSCLC group, as seen in the testing cohort, when compared with Healthy controls. The Kaplan-Meier survival analysis for the localized stage I and IIA cases in the validation cohort, conducted similarly to Fig 3b for the testing cohort, demonstrated a similar survival predicting trend (Fig 4b). Since neither testing nor validation cohorts were involved in the determination of values of the canonical score, by collectively examining all lymph node and metastasis negative cases in both cohorts, the resulting Kaplan-Meier survival predicting capability by the threshold (established by the testing cohort) was enhanced significantly as shown by the insert in Fig 4b.
By combining testing and validation cohorts, and re-calculating the M+SE threshold using all the Prior-to-Dx cases from both cohorts, the resulting Kaplan-Meier survival predictions for all stage I and IIA cases, with patient survival status presented in Fig 5a, remained to be significant (Fig 5b) as that seen in Fig 4b. More clinically relevant, Fig 5c shows that for these cases statistically significant Kaplan-Meier survival rates can be predicted using this threshold according to the date of patients Time-of-Dx, with detailed statistical parameters listed in Fig 5c.
DISCUSSION AND CONCLUSION
In this work, by measuring values of serum MRS metabolomic profiles of samples collected from NSCLC patients both prior-to- and at the time-of -Dx, we demonstrate that LuCa metabolomics measured from sera has the potential to be developed into sensitive and specific profiles that may be implemented as a LuCa screening tool for disease early detections. Furthermore, serum MRS metabolomic profiles, reflecting the biological activities, present thresholds based on those, NSCLC patient survival status can be predicted that may assist to guide clinical strategies for treatment decisions.Acknowledgements
NIH grants CA141139 and the A.A. Martinos Center for Biomedical Imaging.References
No reference found.Figures

Fig 1. Native
sera (10ul) MRS measured with HRMAS. Spectra present as group averages with
standard deviations for training and testing cohorts.
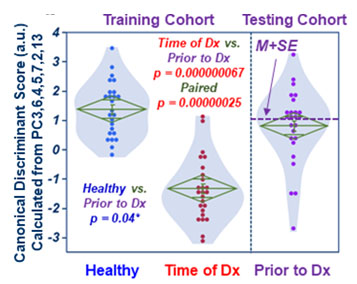
Fig 2. Canonical scores calculated
with training cohort place testing cohort (Prior-to-Dx) between Healthy and
Time-of-Dx.

Fig 3. Kaplan-Meier
survival analysis. a). Mean plus one
standard error (M+SE) of the canonical score differences between Time-of-Dx and
Prior-to-Dx indicates significantly better survival rate from their time of NSCLC
diagnoses for patients with score differences higher than M+SE. b). For Stage I and IIA, patient survivals
can be predicted from the date of their Prior-to-Dx, if their score values are
higher than the M+SE threshold, calculated from the testing cohort (dashed line
in Fig 2).

Fig 4. Validation cohort. a). Validation cohort scores follow those in the training and
testing cohorts. b). The
Kaplan-Meier survival analysis for the Stage I and IIA cases in the validation
cohort, show a similar survival predicting trend. Collectively examining all Stage
I and IIA cases in both cohorts, the resulting Kaplan-Meier survival predicting
capability by the threshold (established by the testing cohort) was enhanced
significantly (insert).

Fig 5. Testing and validation combination. a). Overall patient survival status with the M+SE
threshold (dashed line) calculated using all the Prior-to-Dx cases from both
cohorts. b). The Kaplan-Meier survival predictions for
all stage I and IIA cases. c).
Significant Kaplan-Meier
survival rates predicted using this threshold according to the date of patients
Time-of-Dx, with detailed statistical parameters listed.