3814
Seq2prospa: translating PyPulseq for low-field imaging
Keerthi Sravan Ravi1,2, Thomas O'Reilly3, John Thomas Vaughan Jr.2, Andrew Webb3, and Sairam Geethanath2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, New York, NY, United States, 3Leiden University Medical Center, Leiden, Netherlands
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, New York, NY, United States, 3Leiden University Medical Center, Leiden, Netherlands
Synopsis
Low-field MRI scanners are more viable in resource-strained regions such as Africa, sacrificing SNR for gains on the economics of purchase, siting and service. Pulse sequences need to be optimized for maximum efficiency on these scanners. This work introduces seq2prospa, a Python-based tool facilitating the conversion of pulse sequences designed using PyPulseq into a low-field spectrometer-friendly format. We perform an in-vitro and in-vivo acquisition experiment to image a structural phantom and a human hand respectively. Data acquired from a 2D GRE sequence designed natively in Prospa is compared with data acquired from a PyPulseq-converted 2D GRE sequence.
Introduction
Low-field MRI scanners are more viable in resource-strained regions such as Africa, where countries have <1 scanner per million people as opposed to the world average of 5.30. When compared with 1.5T and 3T scanners, low-field scanners sacrifice SNR for gains on the economics of purchase, siting and service [1]. Pulse sequences need to be optimized for these scanners. Conventionally, designing MR pulse sequences has required knowledge of vendor-specific pulse sequence development (PSD) environments. Currently, to the best of our knowledge, there are no open-source, vendor neutral, tools that enable rapid prototyping and debugging of PSD for low-field MR consoles. PyPulseq [2, 3] has enabled rapid, open-source vendor-neutral, zero-footprint, collaborative PSD. Currently, pulse sequences designed using PyPulseq can be executed on Siemens, GE or Bruker hardware with the aid of vendor-specific interpreters. This work introduces seq2prospa - a Python-based extension to PyPulseq. Seq2prospa is aimed at facilitating the conversion of pulse sequences designed using PyPulseq into a Prospa-friendly format to run sequences on a low-field scanner [4]. Prospa is the software package utilized to execute pulse sequences on Magritek’s Kea2 spectrometer. Currently, seq2prospa supports converting a 2D Gradient Recalled Echo (GRE) sequence. We perform an in-vitro and in-vivo acquisition experiment to image a structural phantom and a human hand respectively. Data acquired from a standard Prospa 2D GRE sequence is compared with data acquired from a PyPulseq-converted 2D GRE sequence. The acquisitions were performed on a 50 mT Halbach array-based MRI scanner [4].Methods
PyPulseq assembles a pulse sequence as a collection of blocks, where each block is a set of simultaneously occurring events. An event is a radiofrequency pulse, spatially varying gradient, time delay or an ADC readout. Prospa constructs a pulse sequence as a collection of successive macros. It switches gradient fields from shim levels to desired amplitudes and leverages time delays to execute combinations of radiofrequency pulses and/or gradients simultaneously. For radiofrequency pulses and ADC readouts, the duration is a parameter that is provided to the function call. Seq2prospa maps blocks in the 2D GRE PyPulseq sequence into successive macros. On reaching the end of a block, seq2prospa switches gradient fields back to shim levels. Gradient fields that are required to maintain their states for longer durations are not modified. An experiment comparing a 2D GRE acquisition programmed natively in Prospa and in PyPulseq was performed. First, a 2D GRE sequence was programmed in PyPulseq and exported as a .seq file. This sequence’s pulse timing diagram was equivalent to that of the 2D GRE sequence designed in Prospa. Subsequently, seq2prospa converted this .seq file into a Prospa-friendly format. The two sequences were executed on a Kea2 spectrometer to perform an in-vitro and an in-vivo acquisition. We imaged a structural phantom and a human hand, and the acquired data was reconstructed for visualization purposes. The structural phantom was tubes of oil spaced 17 mm apart. Data were acquired on a 50 mT Halbach array-based MRI scanner [4]. A 15 cm diameter, 15 cm long solenoid was used as a transmit/receive coil. The following acquisition parameters were used for phantom imaging using the standard Prospa and the PyPulseq-converted GRE sequences: field of view=192 x 192mm, TE/TR=6.5/150 ms and acquisition bandwidth=40 kHz. For hand imaging, the acquisition parameters used were: field of view=256 x 256 mm, TE/TR=6.5/100 ms and acquisition bandwidth=40 kHz. SNR values were measured on the in-vitro data for quantitative comparison of the standard Prospa and PyPulseq-converted sequences. SNR was calculated by computing the ratio of the mean signal over a single tube in the center of the image to the standard deviation of background.Results
The sequence of macro calls in the converted 2D GRE sequence was identical to the pulse sequence originally developed in Prospa. Figure 2 presents the pulse timing diagrams of the 2D GRE sequence from PyPulseq and Prospa. Figures 3 and 4 show image reconstructions of data acquired imaging a structural phantom and a human hand respectively. For the structural phantom, the SNR values for the standard Prospa and the PyPulseq-converted acquisitions were 32 dB and 34 dB respectively.Discussion and conclusion
In this work, we introduced seq2prospa. We demonstrated the first open-source, vendor-neutral, zero-footprint pulse sequence implementation on a low-field scanner through in-vitro and in-vivo experiments. The ability to execute arbitrary Python code in a web browser using Google’s Colab tool enables PyPulseq to run in a web browser. This will allow users to rapidly prototype and debug pulse sequences for low-field MRI systems. Future work involves validating more sequences. In conclusion, this work introduces seq2prospa - an open-source tool that allows users to execute pulse sequences on Magritek’s Kea2 spectrometers, thus enabling data acquisition on low-field systems.Acknowledgements
This study was funded [in part] by the Seed Grant Program for MR Studies and the Technical Development Grant Program for MR Studies of the Zuckerman Mind Brain Behavior Institute at Columbia University and Columbia MR Research Center site.References
- Geethanath, Sairam, and John Thomas Vaughan Jr. "Accessible magnetic resonance imaging: A review." Journal of Magnetic Resonance Imaging 49.7 (2019): e65-e77
- .Ravi, Keerthi Sravan, et al. "Pulseq-Graphical Programming Interface: Open source visual environment for prototyping pulse sequences and integrated magnetic resonance imaging algorithm development." Magnetic resonance imaging 52 (2018): 9-15.
- Ravi, Keerthi Sravan, Sairam Geethanath, and John Thomas Vaughan. "PyPulseq: A Python Package for MRI Pulse Sequence Design." Journal of Open Source Software 4.42 (2019): 1725.
- O’Reilly, Thomas, et al. "In vivo 3D brain and extremity MRI at 50 mT using a permanent magnet Halbach array." Magnetic resonance in medicine 85.1 (2020): 495-505.
Figures
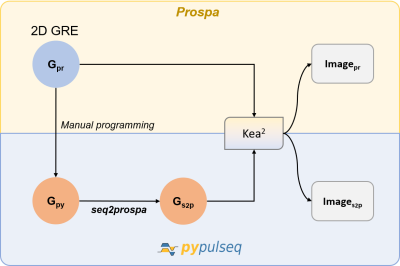
Figure 1. Workflow for converting a 2D Gradient Recalled Echo (GRE) sequence. A 2D GRE sequence designed in Prospa (Gpr) was first manually programmed in PyPulseq (Gpy). Next, this PyPulseq 2D GRE was converted to a Prospa-friendly format (Gs2p) using seq2prospa. Finally, the two 2D GRE sequences (Gpr, Gs2p) were executed on a Kea2 spectrometer to image a human hand and a structural phantom. The acquired images (Ipr, Is2p) were reconstructed and visualized.

Figure 2. PyPulseq and Prospa pulse timing diagrams of the 2D Gradient Recalled Echo (GRE) sequence demonstrated in this work. A 2D GRE sequence was programmed in PyPulseq (Gpy), identical to the 2D GRE sequence developed in Prospa (Gpr). Next, this PyPusleq sequence was converted to a Prospa-friendly format (Gs2p) using seq2prospa. (a, b) Timing diagram of Gpy pulse sequence (c) Timing diagram of Gpr pulse sequence.

Figure 3. Image reconstructions of imaging the structural phantom using the standard Prospa and PyPulseq-converted sequences. The SNR values measured from the standard Prospa acquisition (a) and the PyPulseq-converted acquisition (b) were 32 dB and 34 dB respectively.

Figure 4. Image reconstructions of imaging a human hand using the PyPulseq-converted sequence. Acquisition was performed on a 50 mT Halbach array-based MRI scanner with the following acquisition parameters: field of view=256 x 256 mm, TE/TR=6.5/100 ms and acquisition bandwidth=40 kHz.