3771
Bridging Open Source Sequence Simulation and Acquisition with py2jemris
Gehua Tong1, Sairam Geethanath2, and John Thomas Vaughan, Jr.2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, Columbia University, New York, NY, United States
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, Columbia University, New York, NY, United States
Synopsis
Open-source sequence development tools are often simulation-focused or acquisition-focused. The ability to simulate and acquire from the same sequence file would help speed up method development. In this work, we present an open-source Python tool, py2jemris, which converts arbitrary Pulseq files to JEMRIS simulation format with a zero-footprint Google Colab notebook. A dual simulation/acquisition experiment using the same sequence file was performed to demonstrate the development pipeline. The conversion and simulation time were recorded and evaluated.
Introduction
Recently, open-source platforms have emerged to make MR sequence design, simulation, and application more accessible1-8. These sequence platforms can be divided into simulation-focused ones, such as JEMRIS6 and MRiLab7, and deployment-focused ones, such as Pulseq1 and TOPPE3. Conversion links between the simulation and deployment formats exist but are often incomplete. For example, JEMRIS-built sequences can be exported as Pulseq files, but there is no reverse conversion from Pulseq to JEMRIS. Being able to simulate arbitrary Pulseq files would enable the sequence designer to test the same sequence file in silico ahead of scanning. In this work, we demonstrated a conversion link from Pulseq to JEMRIS along with a sequence simulation pipeline to facilitate prototyping new MR sequences.Methods
py2jemris: The py2jemris toolbox converts Pulseq sequences, phantoms, and Tx/Rx maps into JEMRIS formats; it also allows command-line control of JEMRIS, data loading, and reconstruction (Figure 1). A pipeline was created for efficient testing: it takes a Pulseq file along with other inputs, converts them, starts a JEMRIS simulation, and outputs Cartesian reconstructed images. The repository9 includes the individual functions as well as a Google Colab notebook capable of taking a Pulseq file upload and performing JEMRIS simulation with zero installation.Seq2xml Validation: Seq2xml converts any seq file into the JEMRIS format, a collection of h5 and xml files, by going through the sequence blocks and adding corresponding RF and gradient waveforms to the xml file. All RF pulses and non-trapezoid gradient waveforms require external storage as h5 files, which are referred to within the xml file. In addition, the conversion of phantoms and B1 maps from numpy arrays into JEMRIS-compatible format was included. Seq2xml was validated by double-conversion of a JEMRIS sequence (Figure 2). While the conversion is not completely reversible due to unrolling of the loops, the equivalence of gre.xml and gre2.xml was validated by comparing sequence plots and simulated images.
Experiments: A custom RF-Spoiled Gradient Echo (RF-SPGR) sequence was put through a dual simulation-acquisition experiment (Figure 3). The phantom consisted of two plastic bottles, one containing water and the other vegetable oil. For simulation, A 2D numerical phantom was constructed using segmentation from a vendor-provided Spin Echo acquisition. T1 and T2 mapping using custom Inversion Recovery Spin Echo and Turbo Spin Echo sequences defined the T1 and T2 for the two compartments (water: T1 = 2700 ms, T2 = 1800 ms; vegetable oil: T1 = 290 ms, T2 = 84 ms). Relative proton density of water to oil was calculated by dividing average ROI signals from the FA = 45 degrees SPGR acquisition by their predicted SPGR signals. Simulation was performed using FA = 70 degrees.
Results
Seq2xml Validation: Both the simulated images and the sequence plot show correspondence between the original (gre.xml) and twice-converted (gre2.xml) sequences. The output images are not exactly identical, which we attribute to numerical errors in the conversion of RF waveforms.Experiments: The SPGR images are shown in Figure 4. Acquired images show a shift in relative signal strength between water and oil bottles as the flip angle varied. Artifacts near the edges of the bottles are apparent in the acquired images while absent in the simulated image, where the boundaries were defined by the more geometrically accurate vender sequence. On the other hand, the simulated image contains signal fluctuation often seen in discrete-phantom simulations. Comparing the simulated and acquired images at FA = 70 deg, we obtained PSNR = 19.12 dB and SSIM = 0.7531.
Discussion
While the simulation did not reproduce the geometrical distortion in the acquisition, it demonstrated the ability of the sequence to encode spatial information and contrast. Using the same sequence file for both simulation and acquisition is valuable in two ways: First, it could save scanner cost by precluding basic errors; second, it could help demonstrate sequence effects using arbitrary numerical phantoms. The low-level, single-file representation of Pulseq enables high degrees of freedom in sequence design. Sequence files that are portable and human-readable as well as capable of being directly simulated and acquired on commercial scanners would be uniquely useful to the open-source MR community.The major limitation of the pipeline is speed. As Figure 5 shows, while the seq2xml conversion time and actual JEMRIS simulation time remain reasonable for typical N values, the pipeline can be unreasonably slow. We attribute this to the need to read many redundant waveforms stored as separate files: for example, in RF-SPGR, each RF pulse with a different phase had to be stored as a separate waveform instead of being defined analytically because the Pulseq format contains no loop or analytical definitions. Future work is needed to increase efficiency, either by inferring or encoding structure from the Pulseq files, or by designating scripts that co-generate Pulseq and JEMRIS files as the unit of sharing in open-source pulse sequence development.
Conclusion
In this work, we demonstrated py2jemris, an open-source interface that enables JEMRIS simulation of arbitrary Pulseq sequences. Double conversion and a dual simulation-acquisition experiment showed the validity and utility of the interface. A zero-footprint demo for uploading and simulating Pulseq sequences was made available as a Google Colab notebook.Acknowledgements
This work was supported by the Seed Grant Program for MR Studies and the Technical Development Grant Program for MR Studies of the Zuckerman Mind Brain Behavior Institute at Columbia University and Columbia MR Research Center site, and was also performed at Zuckerman Mind Brain Behavior Institute MRI Platform, a shared resource and Columbia MR Research Center site.References
- Layton KJ, Kroboth S, Jia F, et al. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn Reson Med. 2017;77(4):1544-1552. doi:10.1002/mrm.262352.
- Ravi KS, Potdar S, Poojar P, et al. Pulseq-Graphical Programming Interface: Open source visual environment for prototyping pulse sequences and integrated magnetic resonance imaging algorithm development. Magn Reson Imaging. 2018;52(February):9-15. doi:10.1016/j.mri.2018.03.0083.
- Nielsen JF, Noll DC. TOPPE: A framework for rapid prototyping of MR pulse sequences. Magn Reson Med. 2018;79(6):3128-3134. doi:10.1002/mrm.269904.
- Jochimsen TH, Von Mengershausen M. ODIN - Object-oriented Development Interface for NMR. J Magn Reson. 2004;170(1):67-78. doi:10.1016/j.jmr.2004.05.0215.
- Magland JF, Li C, Langham MC, Wehrli FW. Pulse sequence programming in a dynamic visual environment: SequenceTree. Magn Reson Med. 2016;75(1):257-265. doi:10.1002/mrm.256406.
- Stöcker T, Vahedipour K, Pflugfelder D, Shah NJ. High-performance computing MRI simulations. Magn Reson Med. 2010;64(1):186-193. doi:10.1002/mrm.224067.
- Liu F, Velikina J V., Block WF, Kijowski R, Samsonov AA. Fast Realistic MRI Simulations Based on Generalized Multi-Pool Exchange Tissue Model. IEEE Trans Med Imaging. 2017;36(2):527-537. doi:10.1109/TMI.2016.26209618.
- Benoit-Cattin H, Collewet G, Belaroussi B, Saint-Jalmes H, Odet C. The SIMRI project: A versatile and interactive MRI simulator. J Magn Reson. 2005;173(1):97-115. doi:10.1016/j.jmr.2004.09.0279.
- imr-framework/py2jemris: Python library for interfacing with JEMRIS. Accessed October 8, 2020. https://github.com/imr-framework/py2jemris
Figures
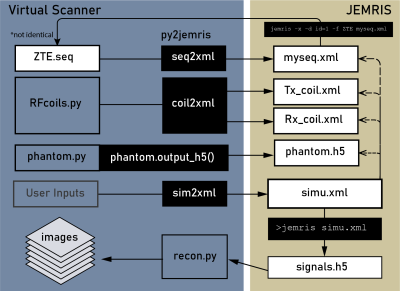
Figure 1: The simulation pipeline enabled by bridging Pulseq to JEMRIS. Sequence, phantom, and B1 map files are converted into JEMRIS format, simulation is performed on the JEMRIS command line, and the resulting data is reconstructed automatically to yield images.
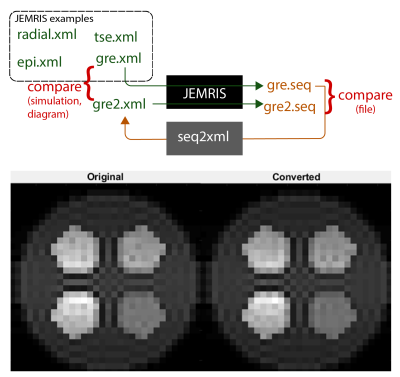
Figure 2: The invariance of the seq2xml conversion script was tested by double-conversion. A JEMRIS-format GRE sequence was outputted in the Pulseq format using JEMRIS. This sequence was converted back to JEMRIS format via seq2xml. The top diagram demonstrates this process. The two JEMRIS-format sequences, (1) the original and (2) the twice-converted, were compared by plotting the sequence waveforms and simulating using a custom 2D phantom, as shown in the bottom plot.
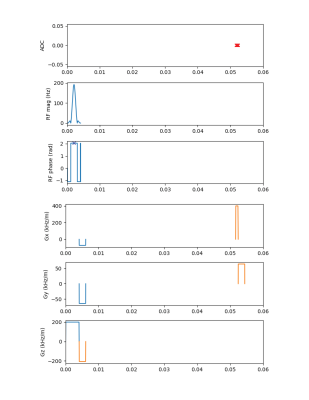
Figure 3: Input waveforms for the first 60 ms are shown for the RF-spoiled SPGR sequence (N = 64, FA = 70 deg, TE = 50 ms, TR = 4500 ms).
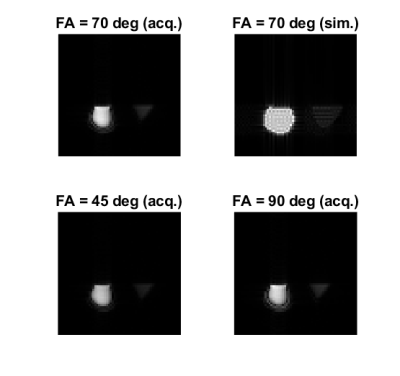
Figure 4: Simulating and acquiring 64 x 64 phantom images from the same sequence file (FOV = 250 mm, slice thickness = 5 mm, TR = 4500 ms, TE = 50 ms). Simulation at FA = 70 degrees was performed while images were acquired with FA = 45, 70, and 90 degrees. At FA = 70 degrees, PSNR = 19.12 dB and SSIM = 0.7531 between the simulated and the acquired images.
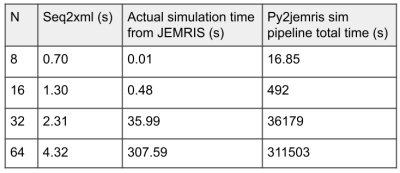
Figure 5. The seq2xml process and simulation pipeline were timed at different matrix sizes N for gradient-spoiled SPGR. Roughly, seq2xml conversion time increases linearly with matrix size while actual simulation time increases quadratically, as expected with 2D phantoms. The total time becomes cumbersome even at smaller N. This was attributed to the need to load each RF waveform separately instead of using analytical definitions. This redundancy was caused by (1) all RF pulses being encoded as arbitrary waveforms by Pulseq and (2) the loopless nature of the Pulseq format.