3768
Reproducibility meets Software Testing: Automatic Tests of Reproducible Publications Using BART1Institute for Diagnostic and Interventional Radiology, University Medical Center Göttingen, Göttingen, Germany, 2Partner Site Göttingen, DZHK (German Centre for Cardiovascular Research), Göttingen, Germany, 3Campus Institute Data Science (CIDAS), University of Göttingen, Göttingen, Germany
Synopsis
Progress in science is only possible if we can trust our current knowledge and build on it. Thus, it is not only important to ensure reproducibility of published results but also to make sure that this is achieved in such a way that building on it is possible. Therefore, we describe a workflow and tool to verify the reproducibility of publications that make use of the BART toolbox. This ensures that published results remain reproducible into the future, even using newer versions of BART.
Introduction
Progress in science is only possible if we can trust our current knowledge and build on it. Reproducibility of published results is therefore of high importance. But as a recent challenge of the ISMRM Reproducible Research Study Group [1] showed, even reproducing a classic publication such as Pruessmann et al.'s CG-SENSE [2] is not entirely straightforward. With the recent push towards reproducibility, publication and preservation of software artifacts is increasingly recognized as a normal and even mandatory part of science.While this is relatively easy to achieve when the software consists of simple analysis scripts which can be distributed with the publication itself, reproducible research becomes much more challenging when the results were obtained with complicated software frameworks as used for advanced image reconstruction, scanner control, or simulation. As a solution it is often proposed to capture the exact version of software environment and archive it (e.g. as a docker container), as this will make it possible to reproduce the results. Still, this is far from ideal: Such frozen environments then contain soon-outdated software environments that will rarely be a able to serve as starting points for future development.
In order to improve this situation for the next generation of scientists, we need to develop a sustainable and integrated workflow for research software development that enables full reproducibility of old results also using new software versions. In this work, we will describe our preliminary experience developing such a workflow using the BART toolbox [3] with reproducible publications that make use of BART as examples.
Methods
For publications that directly include source code and data as supplementary material, reproduction can often be as easy as executing a script. However, in general, a lot more effort is needed, such as: downloading the data (if hosted externally), installation of additional software including all of its dependencies, pre-processing of data, or running multiple reconstruction steps “by hand”.Especially the installation of software might be challenging, if that software is proprietary, depends on complicated our out-dated libraries, old versions of programming languages, or for other reasons.
An additional challenge when working with real data is the size and complexity, and therefore the reconstruction time of some current datasets and methods: While the code for some publications runs relatively quickly (on the order of minutes), reproducing all results of some advanced model-based reconstruction methods can take more than 10 days on a regular computer system.
We based our worklfow on the BART toolbox which was written with longevity in mind and on reconstruction scripts using the Bash shell [4]. While this approach may not be suitable for everybody, we hope that the general concepts can be useful to others as well. Here, we focus on reproducing image reconstruction methods, and therefore only check the reproducibility of reconstruction results in array form. For these, we use the normalized root-mean-square error (NRMSE), compared to a given tolerance, as the criterion for successful reproduction.
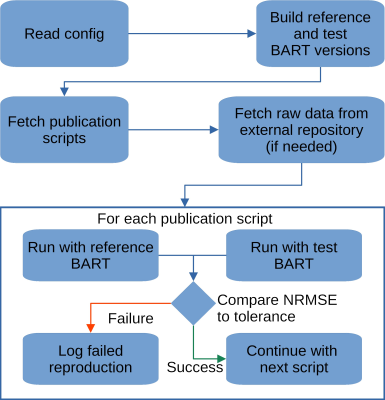
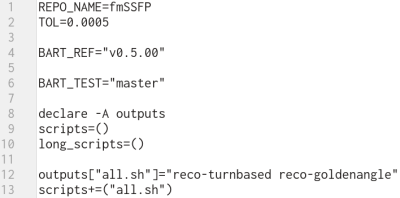
Our reproducibility tool is written in GNU Bash as Bash allows easy calls to arbitrary other programs. It then performs the steps outined in Figure 1. The tools ensures that the original version of BART can still be built and that its results match the current development version. An example of a configuration file can be seen in Figure 2. An overview over the reproducible papers of our group can be seen in Figure 3.
Results
Using a flexible Bash-based approach together with BART, we can ensure that publications stay reproducible even when using newer versions of BART. This is achieved using automatic tests that compare against a reference version of BART.An example illustrating a difficulty when reproducing published results with newer version of a software is the following: A paper about a gradient delay correction method published by our group used BART to calculate a golden-angle trajectory. When the paper was written, the code to calculate the golden ratio used the single-precision floating-point square root function ‘sqrtf’. Because this needlessly uses lower precision, this was later changed to use the double-precision floating point square root ‘sqrt’. However, since this slightly changes the numerical values of the trajectory, the reconstructions are no longer within 0.2% NRMSE (the chose tolerance) of the results using the old code.
Such issues highlight the difficulty of ensuring reproducibility across software versions. Therefore, without automated tests, even correct changes or fixes to a reconstruction software will inevitably lead to the loss of reproducibility over time.
Conclusion and Outlook
In this work, we described workflow and tools to ensure continuing reproducibility of old published result with new versions of BART. These tests simultaneously serve as additional quality control tests in the development of BART. We currently test the reproducibility of our publications periodically after important changes. In the future, we plan to automate runs such tests at regular intervals and before each new release.Beyond BART, we hope that our experiences help other groups to develop a sustainable workflow for reproducibility and software development.
Acknowledgements
Funded in part by NIH under grant U24EB029240 and by the DZHK (German Centre for Cardiovascular Research). We gratefully acknowledge the support of the NVIDIA Corporation with the donation of one NVIDIA TITAN Xp GPU for this research.References
[1]: Maier O et al.: "CG‐SENSE revisited: Results from the first ISMRM reproducibility challenge", Magn. Reson. Med. 2020; 00:1-19. DOI: 10.1002/mrm.28569
[2]: Pruessmann KP et al.: "Advances in sensitivity encoding with arbitrary k-space trajectories", Magn. Reson. Med. 2001; 46:638-651.
[3]: Uecker M et al.: BART 0.6.00 "Toolbox for Computational Magnetic Resonance Imaging", DOI:10.5281/zenodo.3934312
[4]: Free Software Foundation: Bash (5.0.3) [Unix shell program], 2019.
[5]: Roeloffs V et al.: "Frequency‐modulated SSFP with radial sampling and subspace reconstruction: A time‐efficient alternative to phase‐cycled bSSFP", Magn. Reson. Med. 2020; 81:1566-1579. DOI: 10.1002/mrm.27505
Figures