3764
MRSDB: A Scalable Multisite Data Library for Clinical and Machine Learning Applications of Magnetic Resonance Spectroscopy1Center for Clinical Spectroscopy, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, United States
Synopsis
This work introduces a mock standardized library of multisite magnetic resonance spectroscopy (MRS) data and functional online application called MRS Database (MRSDB) for the secure collection, processing, and sharing of that data. The goal of this platform is to enhance global collaboration, improve objective diagnostics in radiology, and facilitate the development of machine learning and artificial intelligence techniques using MRS.
Introduction
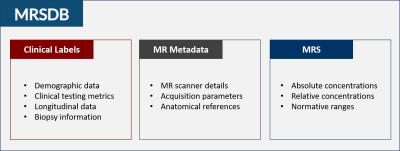
Magnetic resonance spectroscopy (MRS) has been a topic of research for several decades and as such, there exist thousands of published MRS datasets. These datasets could be consolidated into a large and standardized data library to enable researchers to train robust machine learning (ML) and artificial intelligence (AI) models, as well as provide clinicians with a comprehensive normative database to compare their diagnostic data against. To create such a data library, certain requirements must be met to keep the data clinically relevant and ensure reported values are comparable across sites. First, data storage infrastructure must keep protected health information (PHI) confidential. Second, data processing must be unified and vendor-agnostic. Finally, data formats need to be standardized across different systems, field strengths, and vendors. This work presents a mock collection of multisite MRS data generated from real data with the relevant associated clinical and technical metadata, as well as a functional web application called MRS Database (MRSDB) to collect, store, process, and share that data (Fig. 1).Methods
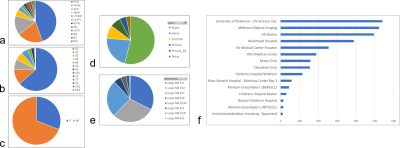
Data Library: The prototype version of MRSDB contains mock data generated from real clinical and research datasets to demonstrate the functionality of MRSDB. The real-life reference library is a unique collection of MRS studies across a multitude of subject matters that include traumatic brain injuries, brain tumors, psychiatric, and metabolic disorders, and includes over 2,000 brain samples using single-voxel spectroscopy and tens of thousands of tissue samples using spectroscopic imaging (Fig. 2). Using these datasets, promising ML applications have been identified, such as modeling brain tissue compositions from metabolic components1, automation of data preprocessing and quantification of spectral signals2, and modeling of behavioral and cognitive scores from brain metabolites in former football players.3,4Standardized Data Format: One goal of this project is to extend the Brain Imaging Data Structure (BIDS) format to encompass MRS5. Once achieved, all current and future data acquired from different sites and systems will be harmonized and standardized using the BIDS format. This will allow the incorporation of BIDS-compatible multimodal data formats into the database. One example is MR imaging, such as structural imaging (T1, T2, FLAIR), diffusion-weighted imaging, and functional MRI, which are often acquired alongside MR spectroscopy. Furthermore, a universal data format will encourage users to contribute their own data to improve and scale the database, thereby enabling the development of more complex and robust ML models over time.
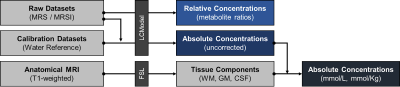
Data Processing, Spectral Quantification, and Brain Tissue Component Extraction: A fully automated and cloud-based back-end service built using Django, a Python-based backend framework, will be implemented to quantify raw MRS data and obtain relative, pseudo-absolute, and absolute concentrations in mmol/L and mmol/Kg (Fig. 3). This service will interface with the LCModel command-line API to compute these values using spectroscopy data, corresponding water reference data, and anatomical volumes. In addition, volume fractions of gray matter, white matter, and cerebrospinal fluid in acquired voxel locations will be extracted from incorporated anatomical information (T1w, T2w, FLAIR) during the data processing pipeline. These components will be added to the database metadata, which will provide additional dimensions for analysis.
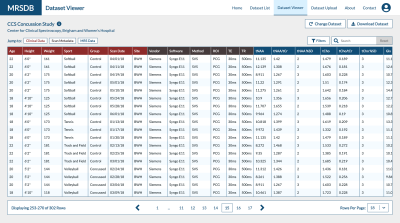
Web Architecture and Resources: The user interface of the database will be a streamlined web application that will facilitate efficient browsing, uploading, and downloading of MRS data (Fig. 4). This platform will be built using ReactJS, a modular and scalable modern web development framework that will allow new features to be added iteratively to accommodate new sites and datasets. The data library will be stored in PostgreSQL, a secure and scalable open-source relational database. The Django back-end service will also handle communications between the database and user interface, which will be secured behind a user-authentication system to prevent unauthorized access of study data and PHI. An example of this interface and functionality can be accessed at https://mrsdb.spectroscopy.org/.
Acknowledgements
No acknowledgement found.References
1. Coello E. et al. Estimation of Brain Tissue Composition and Voxel Location in MR Spectroscopy Using Neural Networks. ISMRM (2019).
2. Louis, M. et al. Quantification of Magnetic Resonance Spectroscopy with unsuppressed Water Signal. ISMRM (2020).
3. Louis, M. et al. Biomarkers for CTE diagnosis in retired NFL players using Machine learning. ISMRM (2018).
4. Charney, M. et al. Cohort Stratification by Clinical Symptoms through Unsupervised Learning Reveals Metabolic and Microstructural Brain Alterations in Former American Football Players. ISMRM (2019).
5. https://forum.mrshub.org/t/bids-for-spectroscopy/83
Figures