3752
A flexible open-source Python package for de-identification of medical images and related data1Flywheel Exchange, Inc, Minneapolis, MN, United States
Synopsis
De-identification of medical data is complex and is a barrier for aggregating heterogeneous data repositories. We have developed a flexible open-source Python package for de-identification of medical images and related data. It is fully featured, supports DICOM and 8 other file types and 9 different field transformations. This package is used in production at Flywheel, Inc. We believe this package can contribute to enforce best standards in the protection of patient identity and privacy while relieving researchers from the burdensome task of developing their own custom tooling.
Introduction
The digital age of medicine has democratized access to a wealth of clinical data that has never before been utilized on a significant scale. One of the critical barriers for aggregating these data repositories is the robust handling of protected health information (PHI) and personally identifiable information (PII)1,2. Redaction of fields containing PHI/PII must occur consistently and accurately in order for data repositories to exist. This task is complicated by varying PHI/PII handling requirements based on regional regulation and local implementation. Adding to this complexity is the need for supporting various data-types (e.g. DICOM, PNG, raw) while applying the same de-identification actions consistently across ancillary data (e.g. JSON, tabular data). Current solutions for de-identification are often restricted to specific domains and do not serve well the extent of medical data at stake in aggregated repositories. In this work, we present a modular yet extensive Python toolkit for de-identification of heterogeneous datasets.Software
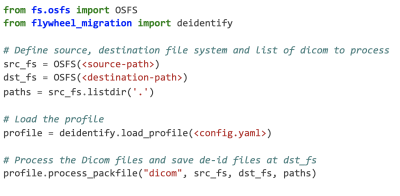
We have developed flywheel-deidentify (https://gitlab.com/flywheel-io/public/migration-toolkit), an open-source Python toolkit that provides a standardized way to redact PHI/PII metadata fields for a variety of file types. The package leverages domain specific file libraries such as pydicom3 and is built around a single human readable YAML de-identification profile that defines the transformations to be applied to all file types. This profile orchestrates the de-identification around three hierarchical layers: 1) A high-level layer that specifies general behavior such as the logging option. 2) A file profile layer that defines the different file types under consideration, their configurations and the file renaming logic. Currently supported file types are: DICOM, PNG, JPEG, TIFF, JSON, XML, Tabular (e.g. CSV, TSV) and key/value text files. 3) A field layer, set for each file profile, that describes the set of transformations applied to the PII/PHI fields. Currently 9 actions are supported: replace-with, remove, set, hash, hashuid (deterministic UUID generation), increment-date, increment-datetime (date and time shift with a predefined increment), jitter (random update of numeric values) and regex-sub (regular expression based substitution).The package relies on a simple API that abstracts away the complexity of the configuration within a YAML profile. For instance, de-identifying DICOM files can be done in a few lines of code as shown in Figure 1. And the process is configured with a simple YAML profile as illustrated for a simple example in Figure 2.
This package is publicly hosted on GitLab under an MIT license and is maintained by developers at Flywheel.io, an informatics platform for biomedical research & collaboration.
Application
This toolkit has been used at scale in production to handle the de-identification of millions of files across different services at Flywheel, including: bulk data ingestion from cloud bucket or local drives, connector in DICOM network, or containerized export process. Originally designed for handling DICOM, its layered architecture makes it easy to extend to other imaging and data types as needed, as well as to integrate within third-party de-identification services. Centered around a single configuration file, the de-identification logic can also easily be shared across users and with local committees for regulatory compliance evaluation. Specifically, particular attention has been given to ensure detection and proper handling of DICOM files with corrupted Value Representations and/or encoding that may forbid PHI/PII redaction by common de-identification tools. Notably, our approach has been tested at scale on DICOM data originating from various manufacturers across multiple modalities.Conclusion
We have developed flywheel-dedentify, an open-source Python package for editing PHI/PII fields in a variety of medical and data file types. To our knowledge, this package represents the first toolkit to provide a large set of field transformations for a substantial number of file types under the same implementation. As medical imaging datasets grow in size and complexity, and regulation becomes more stringent, we believe that this package could contribute to enforce best standards in the protection of patient identity and privacy while relieving researchers from the burdensome task of developing their own custom tooling.Acknowledgements
No acknowledgement found.References
[1] M. Kayaalp, “Patient Privacy in the Era of Big Data,” Balk. Med. J., vol. 35, no. 1, pp. 8–17, Jan. 2018, doi: 10.4274/balkanmedj.2017.0966.
[2] A. Goben and R. J. Sandusky, “Open data repositories: Current risks and opportunities | Goben | College & Research Libraries News,” doi: https://doi.org/10.5860/crln.81.1.62.
[3] Darcy Mason et al., pydicom/pydicom: pydicom 2.1.2. Zenodo, 2020.