3747
Synthetic MRI-assisted Multi-Wavelet Segmentation Framework for Organs-at-Risk Delineation on CT for Radiotherapy Planning1Division of Radiotherapy and Imaging, The Institute of Cancer Research, London, United Kingdom, 2Gynaecological Unit, The Royal Marsden Hospital, London, United Kingdom, 3Department of Radiology, The Royal Marsden Hospital, London, United Kingdom
Synopsis
Radiotherapy (RT) is a cornerstone treatment for cervical cancer. Accurate delineation of organs-at-risk (OARs) is critical to prevent radiation toxicity to healthy organs surrounding the tumour. However, OARs delineation is currently performed manually by clinicians which is labour- and time-intensive. Deep-learning-based algorithms have shown potential in automating this task. This study developed and evaluated a novel framework to incorporate the superior soft-tissue contrast of MRI as well as multi-wavelet image decompositions for improved OARs segmentation on CT images. The proposed framework appears to be a promising addition to the cervical cancer treatment workflow.
Background
In conventional radiotherapy (RT), organs-at-risk (OARs) and target volumes are manually delineated by radiation oncologists on computed tomography (CT) scans which is time-consuming1. Magnetic resonance imaging (MRI) offers excellent soft-tissue contrast and it is often used alongside CT for OARs contouring. Deep learning (DL) approaches have gained popularity to facilitate potential automated OARs and tumour delineation. UNet, is a popular DL architecture used for medical image segmentation2. It uses simplistic max-pooling and up-sampling operations to encode and decode image features during training, which demonstrate information loss3. Wavelets are a more sophisticated means of image restoration/compression, preserving important features4. Despite the recent surge of DL research publications, there is still a need for generalisable and robust segmentation algorithms. This study proposes a novel framework to automate OARs segmentation on CT scans of patients with cervical cancer using generative adversarial network (GAN)-produced synthetic MRI (sMRI) and multi-wavelet UNet (MWUNet).Purpose
Develop and evaluate a deep learning framework for OARs delineation on CT using sMRI and a multi-wavelet UNet to minimise the need for manual contouring by radiation oncologists.Methods
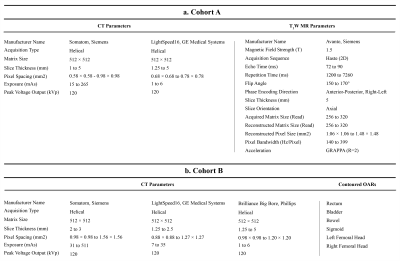
Patient Population and Imaging ProtocolsInstitutional review board approval was obtained for this retrospective study. Two cohorts were identified for MRI synthesis and OARs segmentation: Cohort A consisted of 22 patients with cervical cancer undergoing routine pre-treatment Abdomen-Pelvis CT scanning and T2-weighted MRI (T2W), and Cohort B included 45 patients with cervical cancer and with expert-labelled pre-treatment CT planning contours of OARs and clinical volumes (Table 1).
Framework Architecture and Training
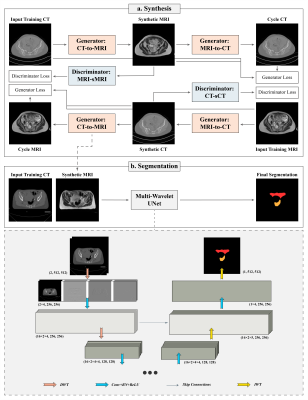
T2W MRI Synthesis from CT (Cohort A)
The T2W MRI images in cohort A were resampled to match the resolution of CT images and subsequently rigidly registered with the corresponding CT slices (loss = Matts mutual information, sampling percentage = 0.5, optimiser = regular step gradient decent). To decrease the dynamic range for input images and enhance model convergence speed, the intensities outside of range -1000 and 1000 HU in CT and 0 and 1500 in T2W MR images were truncated (MR intensities greater than 1500 were present in only 5 patients and 1% of axial slices overall). Subsequently, the images were normalised to values between -1.0 and 1.0. A Cycle-Consistent GAN model (Cycle-GAN) consisting of two generators with 9 residual units (Tanh final layer activation) and two patch-GAN discriminators was developed to facilitate CT-to-MR and MR-to-CT translations5 (Figure 1). The mean absolute error (MAE) and mean squared error (MSE) were chosen as generator and discriminator losses respectively, and they were minimised by the Adam optimiser (learning rate = 0.0002). The network was trained on 16 and evaluated on 6 test patients.
OARs Segmentation (Cohort B)
The 2D axial CT slices and the corresponding contours for rectum, bladder, bowel, sigmoid, left and right femoral heads were extracted from cohort B. The CT intensity values out of the range -500 and 1000 HU were truncated to shorten the dynamic range and normalised to intensities between 0 and 1. A MWUNet was designed such that the max-pooling and up-sampling operations in the baseline UNet2 were replaced with discrete (DWT) and inverse (IWT) Haar wavelet transforms respectively. The trained weights from the Cycle-GAN model were used to generate sMRI from CT images to establish dual-channel input for segmentation training (Figure 1). The proposed framework was trained on 33, validated on 5 and tested on 7 unseen patients respectively using a Nvidia RTX6000 GPU (Santa Clara, California, United States) (loss = average OARs Dice, optimiser = Adam with learning rate 0.0001). Subsequently, the segmentation performance of the proposed approach was compared with MWUNet and UNet models trained solely based on acquired CT images.
Results
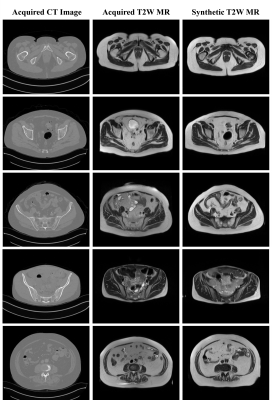
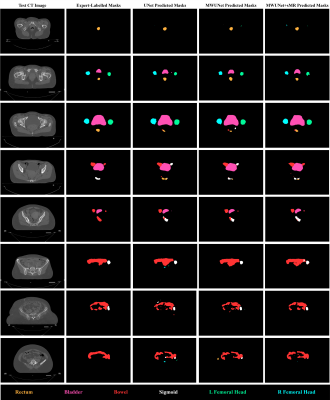
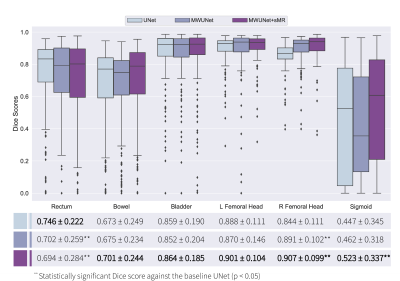
The preliminary results suggested that the proposed framework generated high resolution synthetic T2W MRI from CT images (Figure 2). Subsequently, sMRI-assisted segmentation on CT images using a MWUNet resulted in improved contour predictions of all OARs except the rectum compared with the baseline UNet and MWUNet models trained solely on acquired CT images (Figures 3 & 4).Discussion and Conclusion
One major limitation in RT planning is the need for manual segmentation of regions of interest (ROIs) on CT scans. MRI offers superior soft-tissue contrast compared with CT. This study proposed a novel framework to perform OARs segmentations on CT images using T2W sMRI and multi-wavelet decompositions. The qualitative and quantitative results revealed that this approach led to improved segmentations against the baseline UNet and MWUNet, showing promise in automating OARs segmentation in treatment planning for patients with cervical cancer. Moreover, our results, in line with similar studies6, highlighted the role of sMRI for improved soft-tissue segmentation performance in the pelvis. Future studies will optimise and evaluate the proposed framework for volumetric synthesis and segmentation of OARs and clinical volumes, comparing the performance with state-of-the-art approaches for multi-organ pelvic segmentation, to add further robustness.Acknowledgements
We acknowledge CRUK and EPSRC support to the Cancer Imaging Centre at ICR and RMH in association with MRC and Department of Health C1060/A10334, C1060/A16464 and NHS funding to the NIHR Biomedical Research Centre and the NIHR Royal Marsden Clinical Research Facility. This report is independent research funded by the National Institute for Health Research. The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the National Institute for Health Research or the Department of Health.References
[1] Chen KQ. Systematic evaluation of atlas-based auto segmentation (ABAS) softwarefor adaptive radiation therapy in cervical cancer. China J Radio Med print. 2015;35(2):111–113.
[2] Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Miccai. 2015; 9351:234-241.
[3] Yu D, Wang H, Chen P, Wei Z. Mixed Pooling for Convolutional Neural Networks. Springer International Publishing, 2014; 364–375.
[4] Daubechies I. The wavelet transform, time-frequency localization and signal analysis. IEEE Transactions on Information Theory. 1990; 36(5):961–1005.
[5] Zhu JY, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycleconsistent adversarial networks. In Proceedings of the IEEE international conference on computer 220 vision. 2017; 2223–2232.
[6] Dong X, Lei Y, Tian S, Wang T, Patel P, Curran WJ, Jani AB, Liu T and Yang X. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network. Radiotherapy and Oncology. 2019; 141:192-199.
Figures