3745
Openly available sMall vEsseL sEgmenTaTion pipelinE (OMELETTE)
Hendrik Mattern1
1Biomedical Magnetic Resonance, Otto-von-Guericke-University Magdeburg, Magdeburg, Germany
1Biomedical Magnetic Resonance, Otto-von-Guericke-University Magdeburg, Magdeburg, Germany
Synopsis
An easy-to-use and Openly available sMall vEsseL sEgmenTa-Tion pipelinE (OMELETTE) was developed and compared to a Frangi-based benchmark segmentation. Segmentations performance was evaluated for 20 high resolution Time-of-Flight angiograms. Qualitative and quantitative comparison showed OMELETTE's superior segmentation performance.
Introduction
With the advancement of ultra-high field MRI, Time-of-Flight angiography with voxel sizes$$$\leq$$$300µm has become available. To analyze these high resolution data quantitatively, automatic vessel segmentation is required as manual segmentation becomes unfeasible for such large datasets and high resolutions. However, automatic small vessel segmentation is challenging, since the vessel-to-background ratios decrease with decreasing vessel diameter.Here a novel Openly available sMall vEsseL sEgmenTaTion pipelinE (OMELETTE) is presented. The goal of OMELETTE is to provide high quality small vessel segmentations while being easy to use. OMELETTE's performance is compared with a Frangi-based benchmark for 20 publicly available, high resolution ToF angiograms acquired at 7T1.
Methods
OMELETTE utilizes Jerman's filter2 to enhance vessels and then performs a novel thresholding approach to generate a binary vessel segmentation. All source code was developed in Python3 and takes advantage of the scikit-image package. The proposed pipeline and further documentation is publicly available at: https://gitlab.com/hmattern/omelette/Similar to the well-known Frangi filter3, the Jerman filter2 computes the vesselness of an image by analyzing the eigenvalues of the hessian matrix computed for each voxel at multiple scales. In contrast to Frangi's approach, the Jerman's vessel response function (used to generate the actual enhancement) is based on the volume ratio of the Hessian matrix eigenvalues, commonly used for diffusion tensor analysis2. To improve the enhancement for vessels with low vessel-to-background ratios, a regularization is utilized (controlled by $$$\tau$$$=0.5..1.0). Therefore, Jerman's filter requires a single parameter, while Frangi's filter has three free parameters ($$$ \alpha, \beta, \gamma$$$).
Commonly the enhancement is converted into a segmentation by applying an empirical threshold which trades of the detection of small vessels and falsely segmented noise. To overcome this lengthy trail-and-error tuning, thresholds in OMELETTE are selected automatically with a 3-class Otsu histogram analysis (returning a low and high threshold). Voxels above the high threshold can be considered vessels with high confidence, while voxels below the low threshold are considered background. To leverage this information and differentiate background and vessels in between the thresholds, hysteresis thresholding is used4. Hysteresis thresholding is a bi-threshold procedure in which voxels above the higher threshold are segmented directly. Voxels above the lower threshold are only segmented if they are connected to voxels above the high threshold. This enforces vessel continuity and reduces segmentation of noise.
To improve small vessel segmentation further, OMELETTE provides a novel iterative thresholding approach with soft-enhancement (inspired by compressed sensing's soft-thresholding). As shown in Fig. 1, the segmentation results are propagated back to the filter enhancement. If a voxel was classified as a vessel, its enhancement is increased by the enhancement factor $$$\kappa$$$. Thus, the enhancement $$$E$$$ in the next iteration $$$i+1$$$ is $$$ E(i+1) = E(i) + E(i)*\kappa*S(i)$$$ with $$$S$$$ as the binary segmentation. This process is repeated $$$N$$$ times and intensities above 1.0 are clipped.
To evaluate the segmentation performance, the publicly available 7T ToF angiograms of the studyforrest dataset1 have been used. Prior to segmentation, the 300µm isotropic data of all 20 subjects were bias field-corrected with N4 (provided by ANTs)5. The commonly used Frangi filter3 and segmentation with a single empirically selected threshold was used as the benchmark pipeline. The following parameters were used: $$$\tau$$$=0.5, $$$\kappa$$$=0.05, 50 iterations (OMELETTE); $$$\alpha$$$=0.5, $$$\beta$$$=0.5 ,$$$\gamma$$$=0.05, threshold=0.05 (benchmark); scales=1.0:0.5:2.5 (both pipelines). For quantitative comparison of sensitivity, specificity, and dice coefficient, ilastik's semi-automatic pixel classification6 was used to generate reference segmentations.
Results
Segmentation performance of OMELETTE and the Frangi-based benchmark was compared quantitatively (see Fig. 2). With respect to the semi-manually segmented reference, the specificity of both pipelines was on par. For the sensitivity and dice coefficient OMELETTE returned 53.2% and 5.4% higher scores than the benchmark, respectively.For qualitative comparison of the segmentation performance, axial, whole brain Maximum Intensity Projections (MIPs) and coronal MIPs over the basal ganglia are shown in Fig. 3 and 4, respectively. In line with the quantitative evaluation, OMELETTE provided better segmentation performance than the benchmark. Further, OMELETTE outperformed the reference segmentation. Hence, even with semi-automatic approaches, small vessel segmentation in high resolution 3D data is challenging.
Discussion
With the combination of Jerman's filter and a novel thresholding approach, OMELETTE outperformed the benchmark quantitatively and qualitatively. Further, the qualitative comparisons showed an advantage of OMELETTE compared to the reference. Although, the reference cannot be considered a ground truth, which represents a bias in the analysis, the quantitative results are still sensible. Previous studies showed that Jerman's filter outperform Frangi's filter for enhancing vessels with low contrast2. Further, the qualitative results show the superior performance of the proposed pipeline.Besides the improved segmentation performance and being openly available (source code, data, and auxiliary tools), OMELETTE is easy to use as the enhancement filter requires tuning of single parameter and thresholds are selected automatically.
In the future, OMELETTE will be compared to further segmentation pipeline7,8 and results for the DRIVE challenge9 (retina vessel segmentation with available ground truth) will be reported.
Conclusion
OMELETTE is a novel, easy to use, publicly available vessel segmentation pipeline which outperforms the Frangi-based benchmark.Acknowledgements
This work was funded by the DFG-MA 9235/1-1.References
- Hanke M, Baumgartner FJ, Ibe P, Kaule FR, Pollmann S, Speck O, Zinke W, Stadler J. A high-resolution 7-Tesla fMRI dataset from complex natural stimulation with an audio movie. Sci. Data. 2014;1(1):140003. Available from: https://www.nature.com/articles/sdata20143 doi: 10.1038/sdata.2014.3 .
- Jerman T, Pernus F, Likar B, Spiclin Z. Enhancement of Vascular Structures in 3D and 2D Angiographic Images. IEEE Transactions on Medical Imaging. 2016;35(9):2107–2118. doi: 10.1109/TMI.2016.2550102 .
- Frangi AF, Niessen WJ, Vincken KL, Viergever MA. Multiscale vessel enhancement filtering [Internet]. In: Wells WM, Colchester A, Delp S, editors. Medical image computing and computer-assisted intervention: MICCAI'98 fist international conference, Cambridge, MA, USA, October 11-13,1998 proceedings, vol. 1496. Berlin: Springer-Verlag; 1998. p. 130–137. (vol. 1496).
- Fraz MM, Basit A, Remagnino P, Hoppe A, Barman SA. Retinal vasculature segmentation by morphological curvature, reconstruction and adapted hysteresis thresholding [Internet]. In: 2011 7th International Conference on Emerging Technologies (ICET 2011): Islamabad, Pakistan, 5 - 6 September 2011. 2011 7th International Conference on Emerging Technologies (ICET); 9/5/2011 - 9/6/2011; Islamabad, Pakistan. Piscataway, NJ: IEEE; 2011. p. 1–6.
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–1320 doi: 10.1109/TMI.2010.2046908 .
- Berg S, Kutra D, Kroeger T et al. ilastik: interactive machine learning for (bio)image analysis. Nat Methods. 2019;16(12):1226–1232 doi: 10.1038/s41592-019-0582-9 .
- Bernier M, Cunnane SC, Whittingstall K. The morphology of the human cerebrovascular system. Hum Brain Mapp. 2018. doi: 10.1002/hbm.24337 .
- Huntenburg JM, Steele CJ, Bazin P-L. Nighres: processing tools for high-resolution neuroimaging. Gigascience. 2018;7(7). doi: 10.1093/gigascience/giy082 .
- https://drive.grand-challenge.org/
Figures
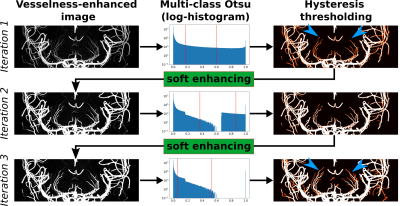
Figure 1: Overview of proposed iterative thresholding. From the vessel-enhanced images two thresholds are selected via multi-class Otsu method. In the hysteresis thresholding all voxels above the higher threshold are segmented (shown in white). Voxel above the lower threshold are only segmented if they are connected to voxels above the high threshold (shown in red). The result of the segmentation is propagated back to the vessel-enhanced image via a novel soft enhancing approach.The entire pipeline is repeated to segment vessels with lower intensity (see blue arrowheads).
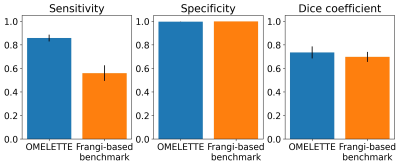
Figure 2: Sensitivity, specificity, and Dice coefficient of OMELETTE and benchmark pipeline for 20 high resolution, publicly available ToF angiographies acquired at 7T. A semi-automatic segmentation (with ilastik) was used as a reference.
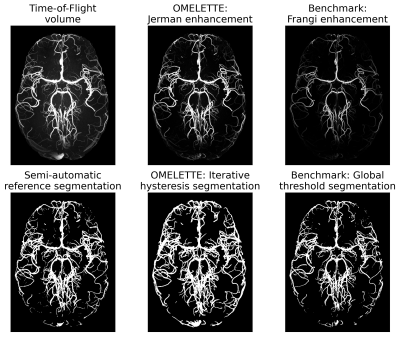
Figure 3: Whole brain comparison of axial maximum intensity projections of the original ToF volume, vesselness-filtering and segmentation output for the proposed OMELETTE and benchmark pipeline. Additionally, the semi-automatically segmented reference is shown. OMELETTE outperforms the benchmark and reference segmentation. Note that all images were normalized, showing that the Jerman enhancement is already more sensitive to small vessel than the Frangi filter.
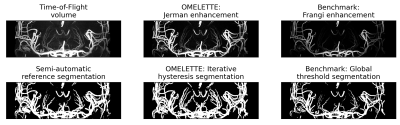
Figure 4: Comparison of coronal maximum intensity projection over the basal ganglia of the original ToF volume, vesselness-filtering and segmentation output for the proposed OMELETTE and benchmark pipeline. Additionally, the semi-automatically segmented reference is shown. OMELETTE enables better segmentation of the lenticulostriate arteries than the benchmark and reference segmentation. Note that all images were normalized.