3736
qVision for the ELGAN-ECHO Study: An MS-qMRI Processing Pipeline Applied to Large-scale, Multi-site, and Multi-vendor Analyses.1Boston University, Boston, MA, United States, 2Boston University Medical Center, Boston, MA, United States, 3University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, MA, United States
Synopsis
Purpose: To describe an integrated, semi-automated image processing pipeline for multispectral qMRI, termed qVision. Methods: Dual-clustering and MS-qMRI python algorithms for the Tri-TSE pulse sequence are automatically calculated and harmonized across a dataset of neuroimaging data from adolescents born extremely preterm. Results: Automated processing is completed in 30 minutes per subject, resulting in high-resolution mappings of T1, T2, PD, and spatial entropy, as well as heavily R1-weighted images of white matter texture via Synthetic-MRI. Conclusion: qVision has been validated on a large-scale, multi-site, and multi-vendor dataset of neuroimaging data, capable of producing a broad spectrum of MS-qMRI outcomes.
Introduction
With the advent of faster, multispectral quantitative MRI (MS-qMRI) pulse sequences, such as triple turbo spin echo (Tri-TSE), there is a critical need for semi-automated, high spatial resolution image processing tools for mapping brain hydration (PD), indirect measures of tissue cellularity and integrity (T1 and T2), and direct metrics of microstructural organization (SE). Whole brain qMRI can help guide developments for neuroprotective and neurorestorative interventions1 as it generates rich information without ionizing radiation. The purpose of this work was to describe an integrated, semi-automatic image processing pipeline for MS-qMRI, termed qVision, and evaluate its utility as part of the multi-site Extremely Low Gestational Age Newborn (ELGAN-ECHO) study2.Methods
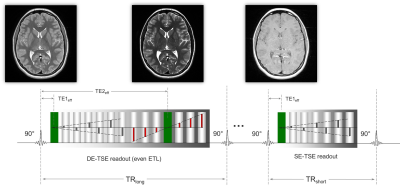
This study was approved by the Institutional Review Boards of the 12 participating institutions of the ELGAN-ECHO study. At 15 years of age, 651 adolescents born extremely preterm (EP) were recruited and returned for follow-up, of which 465 consented for MRI. A 3T MRI protocol using a triple weighting --DA1 = PD-weighted, DA2 = T2-weighted, and DA3 = T1-weighted-- acquisition, termed the triple turbo spin echo pulse sequence (Tri-TSE, Figure 1). The concatenated long repetition time dual echo turbo spin echo (DE-TSE) and short repetition time single echo turbo spin echo (SE-TSE) sequences were implemented with identical scan geometry and TEeff1, 2 of 12ms and 102ms, TRlong of 10s, and TRshort of 0.5s.The image processing pipeline (qVision) consists of dual-clustering segmentation and mapping algorithms programmed in Python 3.7 with the Enthought Deployment Manager. Two preparation steps are required and performed with Fiji: 1) manual editing of the segmented intracranial matter (ICM) segment and 2) manual delineation of the cerebellum’s superior aspect. Multispectral qMRI algorithms were derived as functions of pulse sequence parameters according to the Bloch equation model of the Tri-TSE, which is applicable across all MRI platforms (Eq. 1-3).
Eq. 1: $$$T_2=cf_3\frac{TE1_{eff}-TE2_{eff}}{ln\left(\frac{DA_2}{DA_1}\right)}$$$
Eq. 2: $$$PD=\frac{cf_1}{C_{coil}}\cdot\frac{DA_1\exp\left(\frac{TE1_{eff}}{T_2}\right)+DA_2\exp\left(\frac{TE2_{eff}}{T_2}\right)}{1-\exp\left({\frac{-\left(TR_{long}-TSEshot_{DE}\right)}{T_1}}\right)}$$$
Eq. 3: $$$T_1=Root_{hybr}\left\{T_1+\frac{TR_{short}}{ln\left(\frac{1-\left(\frac{DA_3}{DA_1}\right)\left[\left(1-\exp\left(\frac{-TR_{long}}{T_1}\right)\right)-cf_2\exp\left(\frac{-\left(TR_{long}-TSEshot_{DE}\right)}{T_1}\right)\right]}{1+cf_2\exp\left(\frac{TSEshot_{SE}}{T_1}\right)}\right)}\right\}$$$
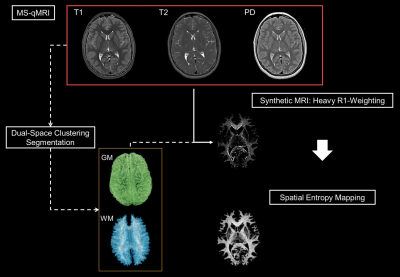
Here, TSEshot_de and TSEshot_se are defined as the product of the echo train length and the echo spacing for the concatenated sequences. Multi-site harmonization was automatically calculated through normalization of mean white matter (WM) qMRI outcomes via three distinct calibration parameters: cf1, cf2 and cf3. Iterative manipulation of the calibration parameters was conducted until inter-scanner discontinuities were minimized. The WM texture latent in PD maps was uncovered via R1-weighted Synthetic-MRI3. Python’s entropy function mapped the spatial entropy of the WM texture to calculate to complexity of the microstructure. For each subject, the PD-T1-T2 qMRI maps and histograms are exported, along with MS-qMRI reports of average tissue parameters for each of the three gross anatomical regions of interest: ICM, cerebrum, and cerebellum.
Results
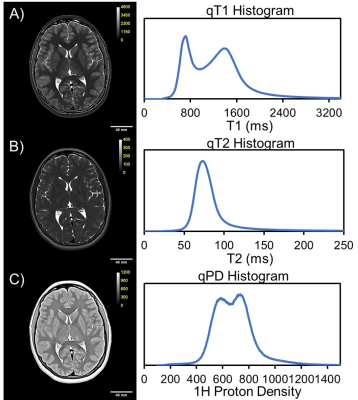
The Tri-TSE is minimalistic and efficient, generating PD-, T2-, and T1-weighted images with a combined scan time of 7:34 minutes. Manual data preparation of each subject is approximately 30min, and is only required once, after which the cerebrum and cerebellum segments are stored for repeated processing in the database. qVision sequentially and automatically processes all subjects in the cohort of interest at 30 minutes per subject (Figure 2). As it relates to the ELGAN-ECHO study, qVision processed a database of 341 subjects unattended within 90 hours. Representative qMRI maps of T1, T2, and PD, and their respective histograms, depict characteristic tissue contrast and expected mean values (Figure 3). Finally, the pipeline generates heavily R1-weighted images and quantitative maps of the white matter’s structural complexity (Figure 2), including qualitative representations of white matter pathways and connections.Discussion and Conclusions
qVision generates a spectrum of high-resolution MS-qMRI data, from measures of hydration and tissue cellularity to structural complexity as measured by spatial entropy. The pipeline has been validated on a large-scale, multi-site, and multi-vendor dataset of neuroimaging data. qVision has several applications, from quantification of dysmaturity states resulting from preterm birth, to innovations in connectomics, and synergizing qMRI data quality in large-scale cohorts. Future work is required to improve automated harmonization via shared, uniform phantom experiments, and standardizing normative values of white matter complexity.Acknowledgements
This work was supported in part by the National Institute of Neurological Disorders and Stroke (5U01NS040069-05 and 2R01NS040069-09), National Institutes of Health Office of the Director (1UG3OD022348-01), and the National Institute of Child Health and Human Development (5P30HD018655-28).References
1. Volpe JJ. Dysmaturation of premature brain: importance, cellular mechanisms, and potential interventions. Pediatric neurology 2019;95:42-66.
2. O'Shea T, Allred E, Dammann O, et al. The ELGAN study of the brain and related disorders in extremely low gestational age newborns. Early human development 2009;85(11):719-725.
3. Jara H. White matter fibrography by synthetic magnetic resonance imaging. Google Patents; 2020.
Figures