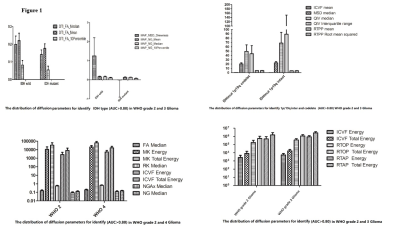
3673
Histogram analysis in prediction of Isocitrate Dehydrogenase Genotype in Gliomas with MRI: The Gaussian versus non-Gaussian Diffusion Models1Dept. of MRI, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 2MR Scientific Marketing, Siemens Healthcare, Shanghai, China, Shanghai, China
Synopsis
The Isocitrate dehydrogenase (IDH) genotyping and epigenetic 1p/19q codeletion as two key molecular markers are included in the glioma WHO 2016 classification. Gaussian or non-Gaussian diffusion models were recently proposed to provide additional microstructure information. In present work, we applied four diffusion models in glioma grading and genotyping, including DTI, DKI, MAP-MRI and NODDI models, which could be acquired within a single scan.
Background and Purpose
According to the IDH and 1p/19q genetic status and prognosis of patients, gliomas could be divided into 3 groups: IDH wild type, IDH mutant with 1p/19q uncodeleted, and IDH mutant with 1p/19q codeleted [1]. Previous studies have proved apparent diffusion coefficient (ADC) and Diffusion tensor imaging (DTI) are useful to prediction of IDH [2,3]. Advanced diffusion models were recently proposed to provide additional microstructure information [4]. This study aimed to evaluate the performance of the 4 diffusion models in glioma genotyping, including Diffusion tensor imaging (DTI), Diffusion kurtosis imaging (DKI), Mean apparent propagator (MAP)-magnetic resonance imaging (MRI), and Neurite orientation dispersion and density imaging (NODDI) models. A single and comprehensive acquisition scheme was used for all the four models. Histogram features were extracted from parameters of these diffusion models and used in genotyping of glioma.Materials and Methods
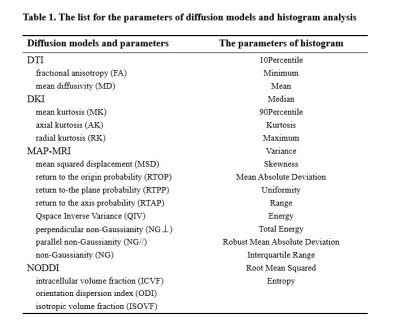
Totally 99 patients were recruited, including grade II (n = 33), grade III (n = 11), or grade IV (n = 54) gliomas. In grade II and III include IDH wild-type gliomas (n = 13), IDH-mutant with 1p/19q uncodeleted (n = 15), and IDH mutant with 1p/19q codeleted (n = 16). All the patients underwent diffusion weighted imaging (DWI) and conventional MRI examinations on a 3T MR scanner (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany) with a 64 channel of head-neck coil. DWI was performed using a spin-echo echo-planar imaging sequence and the parameters were: FOV = 220 × 220 mm2, slice thickness = 2.0 mm, slice number = 66, TR/TE = 3700/72 ms, in-plane acceleration factor = 2, slice acceleration factor = 2, diffusion time δ/Δ = 15.9/35.0 ms, two baseline (b=0) and 98 DWI data with different diffusion directions and b-values were acquired using Cartesian-grid-sampling scheme and bmax= 3000 s/mm2. The DTI, DKI, MAP, and NODDI parameters were calculated using an in-house developed post-processing software called NeuDiLab, which is based on an open-resource tool DIPY (Diffusion Imaging In Python, http://nipy.org/dipy). The calculated parameters are listed in Table 1. The volume-of-interest (VOI) was manually drawn around the entire tumor and peritumoral edema on axial T2-Dark fluid images. The VOI were spatially transfer to the corresponding diffusion parameters by rigid transformation between T2 and baseline diffusion image. The performance of the feature was evaluated using receiver operating characteristic (ROC) curve analysis and the area under the ROC curve (AUC). The diffusion parameters of AUC >0.80 were accept.Results
For IDH type differentiation in WHO grade II and III gliomas, the FA, MSD skewness and NG values from DTI and MAP models outperformed the other parameters of DKI or NODDI. When 1p/19q status were considered in IDH mutation of WHO grade II and III gliomas, the ICVF, MSD, QIV, and RTPP showed outstanding performance. For glioma grading, no diffusion parameter showed AUC>0.80 in differentiating between grade III and IV. To differentiating between grade III and IV, ICVF, RTOP and RTAP showed relatively advantage. FA, MK, RK, ICVF, NGax and NG are more important parameters in differentiating grade III and IV. The distribution of mean and SD of useful parameters were shown as Figure 1.Discussion
MAP diffusion model outperformed the other 3 models in differentiating between genotype groups and different grading groups. Our research founded high sensitivity of NODDI and MAP parameters in differentiating groups with different 1p/19q gene status and IDH mutation, which is different to the finding in Matteo Figini’s research [2]. Histogram analysis offer additional quantitative information of the microstructure information.Conclusion
MAP as an advanced diffusion model has more advantages than other models in prediction of glioma Genotype. Histogram analysis is a useful method in quantitative analysis of diffusion parameters and shows a great potential in glioma research.Acknowledgements
noneReferences
1. Chen, Ricky, et al. "Glioma Subclassifications and Their Clinical Significance." Neurotherapeutics 14.2(2017):1-14.
2. Figini, Matteo et al. “Prediction of Isocitrate Dehydrogenase Genotype in Brain Gliomas with MRI: Single-Shell versus Multishell Diffusion Models.” Radiology vol. 289,3 (2018): 788-796.
3. Maynard, John, et al. "World Health Organization Grade II/III Glioma Molecular Status: Prediction by MRI Morphologic Features and Apparent Diffusion Coefficient." Radiology 296.1(2020):191832.
4. Avram, Alexandru V., et al, “Clinical feasibility of using mean apparent propagator (MAP) MRI to characterize brain tissue microstructure”, Neuroimage 127,(2016) 422–434