3656
Development of in vivo human brain DTI-MRE: Optimization of experimental parameters1Richard and Loan Hill Department of Bioengineering, University of Illinois at Chicago, Chicago, IL, United States, 2Beckman Institute, University of Illinois at Urbana-Champaign, Urbana-Champaign, IL, United States
Synopsis
Simultaneous acquisition of diffusion tensor imaging (DTI) and magnetic resonance elastography (MRE) has been proven feasible in a preliminary study of in vivo human brain. However, the experimental parameters have to be optimized in order to prevent mutual interferences of DTI and MRE acquisitions. We identified in simulations three experimental parameter sets for in vivo human brain DTI-MRE that we classify as good, moderate and poor with regard to optimization and present a pilot study using these parameter sets. The experimental results verify the simulations as we found the best performance of DTI-MRE for the good parameters set.
INTRODUCTION:
Magnetic Resonance Elastography (MRE) and Diffusion Tensor Imaging (DTI) are two non-invasive MRI techniques that use the phase and magnitude of the complex-valued MRI signal, respectively. The simultaneous acquisition of MRE and DTI is beneficial by reducing the scanning time by a factor of ~2 and providing immediately co-registered images. We previously have shown the feasibility of simultaneous acquisition of DTI and MRE, which we name DTI-MRE, on in vivo human brain.1 However, a limitation of DTI-MRE is the tissue vibration might induce signal loss on the magnitude images due to the intra-voxel phase dispersion (IVPD) for certain experimental parameters. Therefore, a careful selection of experimental parameters is needed for DTI and MRE acquisitions not to interfere with each other. We identified optimized experimental parameter sets in simulations for in vivo human brain DTI-MRE.1 In the presented preliminary study we compare the performance of three experimental parameter sets we classify as good, moderate, and poor.METHODS:
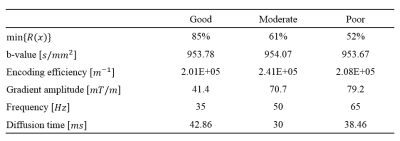
The conditions for finding the three experimental parameter sets are a b-value within the range of 950-1000 s/mm2, an encoding efficiency of vibration above 1.5x105 rad/m, vibration frequencies within range of 20-70 Hz, and a separation time between motion encoding gradients (MEG) selected to be integer multiples of vibration periods.3 The three parameter sets were identified based on the extent of vibration-induced signal loss min{R(x)}, which is listed in Table 1 and was calculated using an equation that was described previously.2The experimental DTI-MRE study on in vivo human brain was approved by the Institutional Review Board (Protocol # 2018-1042) at the University of Illinois at Chicago. The study was conducted on a 3T human scanner (Prisma, Siemens). Previously modified single-shot, spin-echo echo planar imaging (SS-SE-EPI) sequence for DTI-MRE was used in this study. The voxel size, number of slices, matrix size, and TR/TE are 3x3x3 mm3, 48, 80x80 and 6000/80 ms, respectively. Standard EPI MRE without diffusion encoding and EPI DTI acquisitions without vibration were acquired for comparison with the DTI-MRE acquisition. Diffusion property maps and the complex shear modulus G* were calculated as previously described.2 The Spearman correlation was determined voxel-wise between DTI-MRE and conventional methods in a central slice excluding fluid-filled ventricles.
RESULTS:
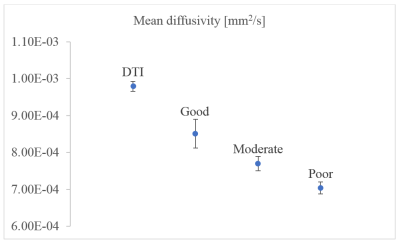
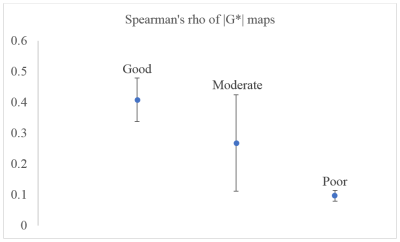
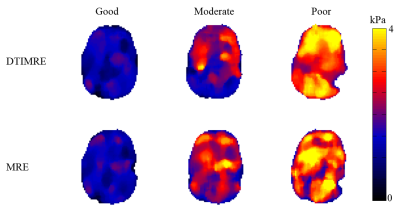
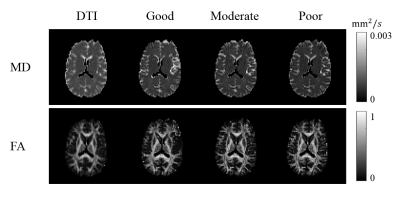
The simulated parameter sets classified as good, moderate, and poor used in our experiments are displayed in the Table 1. Figure 1 shows the reconstructed mean diffusivities (MD) of one subject. The MD value of the good parameter set in DTI-MRE is comparable to values from conventional DTI, and the difference in MD values decreases from good to moderate and poor. Figure 2 displays the Spearman’s correlation coefficients on absolute stiffness maps in a central brain slice of the same subject between DTI-MRE and conventional MRE. A similar correlation trend is observed that confirms the simulations. A direct comparison of reconstructed MRE and DTI parameter maps with DTI-MRE is shown in figs. 3 and 4.DISCUSSION:
Our preliminary results suggest that the simulations provide DTI-MRE experimental parameters optimized for best performance of the new technique. We will conduct a study with a larger sample size in the next months to confirm these observations. DTI-MRE has the potential to increase the clinical acceptance of MRE and DTI by providing fast acquisitions and tissue mechanical and diffusion property maps that are immediately co-registered.Acknowledgements
This work was supported by the NIH NIBIB under grant R21EB026238, Adding MRE to DTI for free. The work represents the views of the authors and not of the NIH.References
1. Lin S, Sutton B, Magin RL, Klatt D: Development of in vivo human brain DTI-MRE. Proceedings of the 28th Annual Meeting of the ISMRM , p. 3325, virtual, 2020.
2. Yin Z, Kearney SP, Magin RL, Klatt D. Concurrent 3D Acquisition of Diffusion Tensor Imaging and Magnetic Resonance Elastography Displacement Data (DTI-MRE): Theory and In Vivo Application. Magnetic Resonance in Medicine 2017; 77(1): 273-284.
3. Yin Z, Magin RL, Klatt D. Simultaneous MR elastography and diffusion acquisitions: Diffusion-MRE (dMRE). Magnetic Resonance in Medicine 2014; 71(5): 1682-1688.
Figures