3575
2D and 3D Spiral for Diffusion Weighted MRI with Hyperpolarized 129Xe1Center for Pulmonary Imaging Research, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2Biomedical Engineering Department, University of Cincinnati, Cincinnati, OH, United States, 3Philips, Cincinnati, OH, United States, 4Department of Pediatrics, University of Cincinnati, Cincinnati, OH, United States, 5Division of Pulmonary Medicine, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
Synopsis
Diffusion-weighted hyperpolarized 129Xe MRI is a validated measure of lung microstructure and can assess changes in alveolar dimensions. These images are commonly acquired via 2D gradient recalled echo (GRE), but 129Xe GRE images suffer from coarse resolution in the slice dimension and breath-hold durations (≤16 s) that may be difficult for pediatric and severely ill subjects. To overcome these limitations, we implemented 2D- and 3D-spiral (FLORET, Fermat looped, orthogonally encoded trajectories) sequences for 129Xe diffusion imaging. These sequences enable either rapid acquisition or high-resolution, isotropic lung coverage and display image quality and ADC accuracy comparable to that of conventional 2D-GRE.
Introduction
Hyperpolarized (HP) 129Xe MRI is a powerful, non-invasive tool to quantify regional lung function and structure1-4. Diffusion-weighted MRI—specifically the 129Xe apparent diffusion coefficient (ADC)—is a validated measure of lung microstructural dimensions, with elevated ADC reflecting alveolar enlargement5,6. Diffusion-weighted HP 129Xe MRI is typically performed with slice-selective 2D-GRE sequences using relatively thick slices to span the entire lung volume within a held breath. Such long breath holds (up to 16 s) can be difficult to perform for pediatric subjects and severely ill patients. Some non-Cartesian methods such as spirals acquire k-space data more efficiently, requiring fewer excitations for diffusion imaging, thus reducing overall scan duration. As such, these methods have been successfully implemented for HP 129Xe of ventilation7,8, but they have yet to be explored for HP 129Xe diffusion imaging. Here we apply 2D and 3D spiral (3D FLORET, Fermat looped, orthogonally encoded trajectories 9,10) for HP 129Xe diffusion imaging and compared the results to those obtained via conventional 2D-GRE.Methods
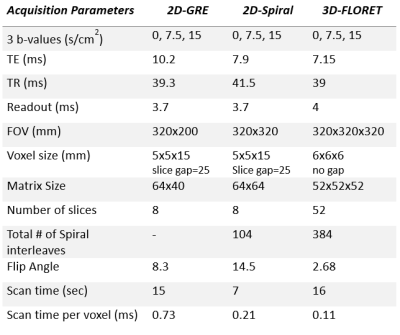
A 2D-spiral multi-slice sequence was combined with bipolar diffusion encoding gradient (diffusion time, Δ=3.5 ms) for multi-b-value diffusion imaging. Additionally, a 3D-spiral sequence, based on the Fermat spiral7,9,10 was also tested for 3D isotropic 129Xe diffusion imaging. In the FLORET sequence, each spiral is projected onto a single cone (between +90° and -90°). One orthogonal set of cones was acquired to fully acquire k-space.To demonstrate the feasibility of these sequences, a healthy male subject (30 years old) was imaged for 129Xe diffusion MRI. Three separate 1L doses of xenon were administered for the comparison between the two spiral sequences and the traditional 2D-GRE sequence. HP 129Xe gas was polarized to 20%, 28% and 29% for each bag (GRE, 2D-spiral, 3D FLORET) on a commercial polarizer (Model 9820, Polarean plc, Durham, NC). Diffusion MR images were acquired on a 3T Philips Achieva scanner with a flexible transmit/receive 129Xe chest coil (Clinical MR Solutions). The relevant acquisition parameters can be found in Table. 1. In short, 3 b-values 0-15 s/cm2 images were acquired with FOV [GRE=320x200 mm2, Spiral=320x320 mm2, FLORET=320x320x320 mm3], Voxel Size [GRE=5x5x15 mm3, Spiral=5x5x15 mm3, FLORET=6x6x6 mm3], 8 slices for the 2D-GRE and 2D-spiral, 52 slices for 3D-FLORET, scan time [GRE=15 s, Spiral=7 s, FLORET=16 s]. All images were reconstructed using Graphical Programming Interface (GPI)11 with further analysis and ADC estimations (linear fit5) performed in MATLAB (Mathworks, Natick, MA). The SNR was measured for all b-value images and normalized to the polarization (P) of the doses recorded by the time of imaging and voxel volume (V) to mitigate the source of variations related to the available HP magnetization and more informative for comparisons.
Results
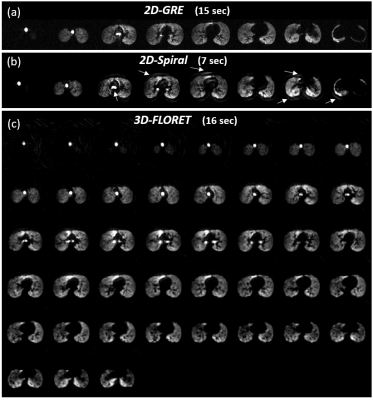
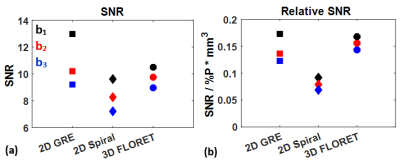
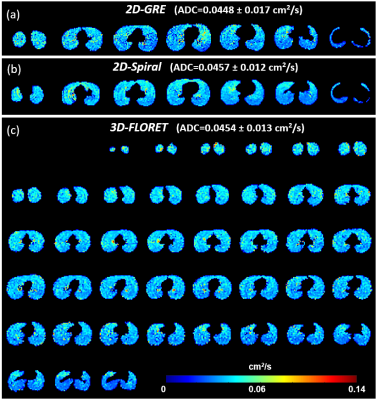
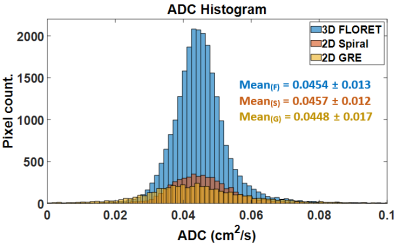
The images of the first b-value (b0) are shown in Figure 1 for the 2D-GRE (a), 2D-spiral (b) and 3D-FLORET (c). Both spiral sequences had image quality broadly comparable to 2D-GRE. However, the 2D sequence displayed some blurring artifacts near high-signal edges, presumably due to a combination of Gibbs ringing (in high SNR in the airways), long readout window, or presence of off-resonance effects. Due to this, relative SNR [SNR/(P·V)] was measured to be slightly higher in the case of 2D-GRE compared to the spiral sequences as shown in Figure 2. Estimated ADC values (Figure 3) were similar for all sequences (ADCGRE=0.0448 ± 0.017, ADC2D-spiral=0.0457 ± 0.012 and ADCFLORET=0.0454 ± 0.013 cm2/s), with no meaningful difference between ADC distributions acquired with either approach (Figure 4).Discussion
2D-spiral and 3D FLORET acquisition approaches were applied to 129Xe diffusion mapping and tested in a healthy volunteer, yielding image quality and ADC values comparable to conventional 2D-GRE encoding. 2D-spiral provided identical lung coverage and resolution but enabled 3-fold faster acquisition. The 2D-spiral is expected to generate higher SNR, because it requires fewer RF excitations, enabling the use of larger flip angles. However, SNR gain was not fully realized, due to noise-like background artifacts, which may be removed through improved image reconstruction. 3D FLORET provide complete lung coverage (no gaps) within the same breath-hold duration (~16 s) with no obvious artifacts, relative to 2D-GRE.Fast 2D spiral sequences have potential applications in subjects who would not be able to perform lengthy breath-holds, younger subjects where compliance may be an issue. The 3D sequence will be advantageous for applications where high resolution and/or full lung coverage is desirable. Future work will involve optimizing the 2D spiral acquisition and reconstruction to further improve image quality.
Conclusion
The use of an efficient 2D and 3D spiral sequences allows for much faster acquisition of 129Xe diffusion imaging or full lung coverage in a single breath-hold maneuvers, enabling shortened breath-hold durations.Acknowledgements
The authors thank the following sources for research funding and support: R44HL123299, R01HL131012 and R01HL143011) and the Cystic Fibrosis Foundation.References
1. Walkup LL, Woods JC JNiB. Translational applications of hyperpolarized 3He and 129Xe. 2014;27:1429-1438.
2. Goodson BM JJoMR. Nuclear magnetic resonance of laser-polarized noble gases in molecules, materials, and organisms. 2002;155:157-216.
3. Ruppert KJ RoPiP. Biomedical imaging with hyperpolarized noble gases. 2014;77:116701.
4. Mugler III JP, Altes TAJJoMRI. Hyperpolarized 129Xe MRI of the human lung. 2013;37:313-331.
5. Kaushik SS, Cleveland ZI, Cofer GP, et al. Diffusion-weighted hyperpolarized 129Xe MRI in healthy volunteers and subjects with chronic obstructive pulmonary disease. Magn Reson Med 2011;65:1154-1165.
6. Thomen RP, Quirk JD, Roach D, Egan‐Rojas T, Ruppert K, Yusen RD, Altes TA, Yablonskiy DA, Woods JC JMrim. Direct comparison of 129Xe diffusion measurements with quantitative histology in human lungs. 2017;77:265-272.
7. Willmering MM, Niedbalski PJ, Wang H, Walkup LL, Robison RK, Pipe JG, Cleveland ZI, Woods JC JMRiM. Improved pulmonary 129Xe ventilation imaging via 3D‐spiral UTE MRI. 2020;84:312-320.
8. Brandon Zanette YF, Samal Munidasa, and Giles Santy Comparison of 3D stack-of-spirals and 2D gradient echo for ventilation mapping using hyperpolarized 129Xe. ISMRM2020.
9. Willmering MM, Robison RK, Wang H, Pipe JG, Woods JC. Implementation of the FLORET UTE sequence for lung imaging. Magn Reson Med 2019;82:1091-1100.
10. Robison RK, Anderson III AG, Pipe JG JMRiM. Three‐dimensional ultrashort echo‐time imaging using a floret trajectory. 2017;78:1038-1049.
11. Zwart NR, Pipe JG JMRiM. Graphical programming interface: A development environment for MRI methods. 2015;74:1449-1460.
Figures