3573
Fully-automated Multi-spectral Pulmonary Registration for Hyperpolarized Noble Gas MRI Using Neural Networks1Medical Biophysics, Robarts Research Institute, Western University, London, ON, Canada, 2School of Biomedical Engineering, Robarts Research Institute, Western University, London, ON, Canada
Synopsis
Co-registered hyperpolarized gas and proton MRI are required to calculate functional lung biomarkers using semi-automated pipelines. Automated registration between different spectral acquisitions is difficult due to differences in contrast and imaging features between different spectral images. Convolutional neural networks create abstract representations of images that may overcome these feature differences. We retrospectively pooled data sets previously registered using a semi-automated pipeline and applied random two-dimensional affine transformations and noise. Neural networks generated inverse transformation matrices from these data to correct the applied transformations. The trained network successfully corrected mis-alignment with an average error of less than one pixel.
Purpose
Hyperpolarized gas MRI (3He and 129Xe) are increasingly relied upon for pathophysiological investigations of lung disease and measuring treatment response. Biomarkers such as ventilation defect percent (VDP)1 or percent high barrier signal2 compare volumes of abnormal lung function to total lung volume. Defects in functional compartments make lung segmentation difficult from hyperpolarized gas images alone, therefore 1H MRI are also acquired to assist segmentation. Small misalignments of images can cause an overestimation of defect volumes, which may be corrected by registration. Established MR processing pipelines co-register gas and 1H data sets using semi-automated landmark registration.1 Semi-automated tools are time consuming and introduce both intra- and inter-user variability. Multi-spectral image sets are challenging to register due to different contrast and features that prevent traditional pattern matching. Due to previous success in multi-spectral segmentation,3 we hypothesized that a neural network could synthesize registration transformations from multi-spectral hyperpolarized gas and 1H images to develop a fully automated image processing pipeline and streamline image acquisition.METHODS:
Participants and Image Acquisition:Data were retrospectively pooled from previous semi-automated registration studies.4-7 1H, 3He and 129Xe MRI were acquired using a 3.0T Discovery MR750 scanner (GE Healthcare, WI) with broadband imaging capabilities. Gas hyperpolarization was performed using a Helispin hyperpolarizer (Polarean, NC) or Polarean 9820 hyperpolarizer (Polarean, NC). 1H MRI were acquired during participant breath-hold of 1.0L nitrogen (N2, Spectra Gases NJ) with a whole-body coil and fast spoiled gradient-recalled-echo sequence (TR/TE/flip=4.7ms/1.2ms/30o, field-of-view=40x40cm2, slice thickness=15mm, number of slices=16-18, bandwidth=24.4kHz, matrix=128x80 padded to 128x128, partial echo=62.5%). 3He MRI were acquired during participant breath-hold of 1.0L of a 40%/60% 3He/N2 mixture using a fast spoiled gradient-recalled-echo sequence (TE/TR/flip= 3.8ms/1.0ms/7o, field-of-view=40x40cm2, slice thickness=15mm, number of slices=16-18 bandwidth=48.8kHz, matrix=128x80 padded to 128x128, partial echo coverage=62.5%). 129Xe were acquired during a participant inhalation of 1.0L 40%/60% 129Xe/4He gas using a coronal plane 3D fast gradient recalled echo sequence (TR/TE/flip= 5.1ms/1.5ms/1.3o, field-of-view=40x40x24cm3, bandwidth=16kHz, matrix=128x128x16-18).
Data Annotation:
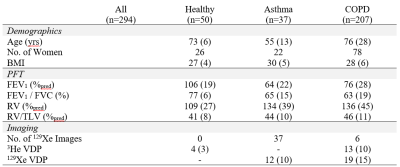
Data were pooled from previous imaging studies in healthy volunteers, participants with asthma, and participants with COPD. Ground-truth registrations were obtained from previously completed semi-automated segmentation pipeline1 output in these studies. Pipeline output included registered image sets but did not include the calculated transform matrices. To generate training data, gas images were transformed using random permutations of scaling (0.8 to 1.2x scale), translation (-16 to +16 pixels), and rotation (-10o to +10o). A subsample of 20 image sets was re-processed to determine the range of translations, scales, and rotations to apply. Corresponding inverse matrices were calculated and normalized on an element-wise basis against the maximum value possible for each element to prevent preferential fitting to the translation. Images were normalized to an intensity range of [0,+1] and Gaussian noise (mean=0, standard deviation = 0.1) was added to prevent perfect overlap.
Network:
Custom neural network architectures were written in python using keras and tensorflow libraries to determine an optimum configuration (Google, CA). Five network architectures were developed to perform registration, compiled with Adam optimization and trained on a GPU (NVIDIA GTX1060Ti, NVidia, CA). Data were split five-fold for cross validation and sampled using a custom keras dataloader from 3D volumes (window = 128x128x1, queue=160, batch=32 per iteration). The network output 2x3 matrices representing a proposed, normalized inverse transform matrix and these were evaluated against the ground-truth inverse transform by mean absolute error.
Results
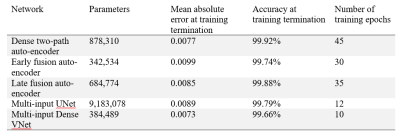
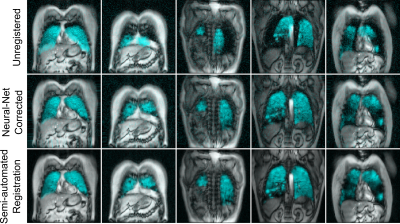
Participant demographics are summarized in Table 1. Analysis of 20 image sets determined the maximum scaling was 0.9x, the maximum translation was eight pixels, and the maximum rotation was 5o and training transformation maximums were set at twice these levels. Registration error is reported in Table 2 for five different network architectures: multi-input UNet, multi-input dense VNet, dense two-path auto-encoder, early fusion auto-encoder and late fusion auto-encoder. The model produced results at a rate of 6ms/slice. Training was stopped when validation loss plateaued. All networks produced results with good visual alignment between gas and 1H images.Discussion
Fully-automated multi-spectral image registration was performed by convolutional neural networks and successfully aligned gas images to the thoracic cavity. The five neural network architectures performed similarly, despite differences in layer layout. A multi-input dense VNet was observed to have the lowest average error, equivalent to a one pixel offset between target and moving images. Interestingly, networks with efficient layouts such as early fusion (342,534 parameters) were nearly as accurate as densely connect networks such as UNet (9,183,078 parameters). This may suggest that affine transformations are easily modeled with fewer variables, even in multi-spectral settings. Densely connected networks such as dense VNet outperformed both early-fusion and late fusion methods.Conclusions
A multi-input, multi-branch UNet successfully generated inverse transforms to perform multi-spectral image registration with average error of 1 pixel offset. Future development may allow more complex transforms (i.e. non-rigid) to be modelled between different spectral data sets and correct for differences in participant inhalation.Acknowledgements
No acknowledgement found.References
1 Kirby, M. Acad Radiol (2012).
2 Niedbalski, P. J. J Appl Physiol(2020).
3 Matheson, A. M. in Proc ISMRM 2020.
4 Kirby, M. Radiology (2010).
5 Svenningsen, S. JMRI (2013).
6 McIntosh, M. in Proc ISMRM 2020.
7 Sheikh, K. J Appl Physiol (2014).
Figures