3515
Automatic segmentation of fetal brain components from MRI using deep learning1Sagol Brain Institute, Tel Aviv Sourasky Medical Center; Israel, Tel Aviv, Israel, 2Sagol School of Neuroscience, Tel Aviv University; Israel, Tel Aviv, Israel, 3School of Computer Science and Engineering, The Hebrew University of Jerusalem, Jerusalem, Israel, Jerusalem, Israel, 4Medical Imaging, Children’s Hospital of Eastern Ontario, University of Ottawa, Ottawa, Canada, Ottawa, ON, Canada, 5Sackler Faculty of Medicine, Tel Aviv University; Israel, Tel Aviv, Israel
Synopsis
Segmentation of the fetal brain into its components is important for quantitative assessment of fetal development. This study proposes a fully automatic method based on deep learning for fetal brain segmentation into six components, including a separation of right and left hemispheres. The method’s performance demonstrated high Dice scores for all brain components and robustness to different contrasts, scan resolutions, gestational age and fetal brain pathologies. Preliminary results demonstrated significant larger ventricle’s volumes and asymmetry in fetuses with ventriculomegaly compared to normal fetuses. The method is suggested to improve fetal assessment and assist radiologists in routine clinical practice.
INTRODUCTION
Fetal MR imaging is increasingly being used as a complementary tool to ultrasound in cases with unclear findings, and shows significant advantages in the assessment of fetal brain malformations. Segmentation of the fetal brain into its components is an essential step for assessing volumetric changes with gestational age (GA), and assessing brain symmetry. While several studies proposed methods for total fetal brain segmentation, only a few further segmented the brain into its components. Manual segmentation was used to create a normal volumetric growth chart of various brain components by GA(1). However, manual segmentation is highly time consuming and requires high expertise. A few studies proposed automatic methods(2, 3). However, they were usually applied on a small cohort, were developed on a single MRI contrast, and did not segment the brain into its two hemispheres. Here we propose a deep learning (DL) method that classifies the brain into six components while separating between the right and left lateral ventricles and the two brain hemispheres. The method is applicable to two MR sequences, several scan resolutions, broad gestational age (GA) range and to both normal and abnormal fetuses. Preliminary results in cases with ventriculomegaly (VM) demonstrate the importance of volumetric assessment over 2D linear measurements, and the separation of right and left hemispheres.Subjects
Fifty-five fetuses (GA 22-37 weeks) were included. Twenty-two were normal developing fetuses and thirty-three were diagnosed with various pathologies including VM, dysmorphic corpus callosum, asymmetry of cerebral hemispheres, and more.MRI Protocols
Data included coronal brain MRI volumes acquired on 1.5T General Electric or 3T Siemens systems and included T2-weighted FRFSE or HASTE sequences, with in-plane resolution of 0.45-1.25mm and slice thickness of 2-5mm.Data annotation
Manual segmentation of brain components was performed using ITK-SNAP software, dividing the fetal brain into five components (labels): (1) Left hemisphere (LH); (2) Right hemisphere (RH); (3) Cerebellum + Brainstem (CB); (4) Lateral Ventricles (LV); (5) CSF - extra axial CSF including the 3rd and 4th ventricles (eCSF). Manual segmentations were approved by an expert neuro-radiologist, and were used as ground truth for model training and testing.Method
The method consists of three stages (Figure 1): (1) fetal brain volume of interest (VOI) detection; (2) fetal brain components segmentation; (3) Post-processing.(1) Fetal brain VOI detection: The fetal brain was first segmented using a 3D two-stage anisotropic U-Net(6), and then cropped using axis-aligned 3D bounding box of the resulting segmentation.
(2) Fetal brain components segmentation: A 2D U-Net consisting of a Resnet34 encoder pre-trained on the ImageNet, similar to(4) was used. The network was trained with the Lovasz loss function(5) for 24 epochs. In the first 12 epochs, only the decoder layers were trained; in the next 12 epochs, both the encoder and decoder layers were trained. Data augmentations including random rotation and intensity inhomogeneity filter(2) were performed. In the online inference phase, test time augmentation(7) was applied where the network segments each slice at various rotation angles, then a majority vote decided the final label of each pixel.
(3) Post-processing: was applied to each slice where either small connected objects are removed, or large connected components, which do not overlap with adjacent slices. Separation between right and left lateral ventricles was done following hemispheres separation by matching the lateral ventricle to its closest hemisphere by checking its contour neighbors (Figure 1 c, d).
Evaluation of the segmentation results
Segmentation network results were evaluated for each brain component on the test set using Dice score, the Hausdorff’s distance and the Asymmetric Surface Distance (ASSD) metrics.Assessment of ventricle volumes and symmetry
Mean values of the ventricular volume (average of right and left ventricles), and ventricular symmetry (ratio between the volume of the large ventricle to the small one) were compared between normal fetuses (n=22) and fetuses with VM (n=12).Algorithm results
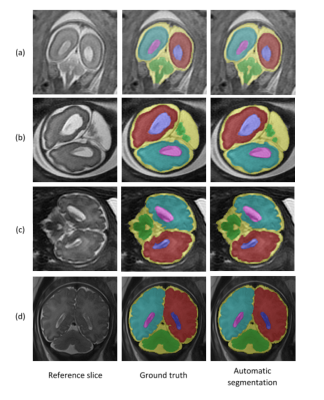
the performance of the algorithm for each component is given in Table 1. The proposed method accurately segmented the fetal brain into six distinct components, and showed robustness to different contrasts, scan resolutions, broad GA range and to both normal and abnormal subjects. Representative results for normal fetuses and fetuses with brain pathologies are presented in Figure 2.Assessment of ventricle volumes and symmetry
Significantly (T-test, p<0.001) smaller ventricle volumes were detected in normal fetuses (2.7cc) compared to fetuses with VM (6.2cc). Significantly (T-test, p=0.01) less asymmetry was detected in normal fetuses (1.26) compared to fetuses with VM (2.05). Preliminary results showed enlarged ventricles in four fetuses, although they were radiologically diagnosed as normal based on 2D manual linear measurements at the level of the atrium.DISCUSSION and CONCLUSION
We propose a fully automated DL based method for fetal brain components segmentation, including a separation of right and left hemispheres. The method was developed and applied in a large clinical cohort. The high performance and preliminary results in cases with VM demonstrate the potential applicability of the proposed method to improve diagnosis and assist radiologists in routine clinical practice. Our current efforts are to optimize the method, increase the cohort in order to create normal fetal brain developmental curves, evaluate the sensitivity of the method to assess brain symmetry, and accurately classify normal vs. abnormal fetuses.Acknowledgements
This work was supported by Kamin grants of the Israel Inovation Authority.References
1. Kyriakopoulou, V. et al, M.A. (2017). Normative biometry of the fetal brain using magnetic resonance imaging. Brain Structure and Function, 222, 2295–2307.
2. Khalili, N., et al.: Automatic brain tissue segmentation in fetal MRI using convolutional neural networks. Magn. Reson. Imaging 64, 7–89 (2019)
3. A. Serag, V. Kyriakopoulou, M. Rutherford, A. Edwards, J. Hajnal, P. Al- jabar, S. Counsell, J. Boardman, D. Rueckert, A multi-channel 4D proba- bilistic atlas of the developing brain: Application to fetuses and neonates, Annals of the British Machine Vision Association 2012 (3) (2012) 1–14.
4. Frid-Adar, M., Ben-Cohen, A., Amer, R., Greenspan, H.: Improving the segmentation of anatomical structures in chest radiographs using U-Net with an ImageNet Pre-trained encoder. In: Stoyanov, D. (ed.) RAMBO/BIA/TIA -2018. LNCS, vol. 11040, pp. 159–168. Springer, Cham (2018).
5. M. Berman, A. Triki, and M. Blaschko. The lovasz-softmax loss: A tractable surrogate for the optimization of the intersection-over-union measure in neural networks. arXiv preprint arXiv:1705.08790, 2017.
6. Dudovitch, G., et al. Deep Learning Automatic Fetal Structures Segmentation in MRI Scans with Few Annotated Datasets. in Proc. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2020.
7. Wang, G., Li, W., Ourselin, S., Vercauteren, T.: Automatic brain tumor segmentation using convolutional neural networks with test-time augmentation. BrainLes 2018, Springer LNCS 11384 (2019) 61–72.
Figures