3510
A comparison of three whole brain segmentation methods for in vivo manganese enhanced MRI in animal models of Alzheimer’s disease1Department of Cybernetics, Czech Technical University, Prague, Czech Republic, 2Department of Neuroimaging, King's College London, London, United Kingdom, 3Biomedical Engineering, Duke University, Durham, NC, United States, 4Georgia Institute of Technology, Atlanta, GA, United States, 5Radiology, Duke University Medical Center, Durham, NC, United States, 6Psychology and Neuroscience, Duke University, Durham, NC, United States, 7Neurology, Duke University, Durham, NC, United States, 8Brain Imaging and Analysis, Duke University Medical School, Durham, NC, United States
Synopsis
Whole mouse brain segmentation is an essential prerequisite for multiple quantitative image analysis tasks and pipelines. In this work, we compare three methods for whole brain segmentation (skull stripping) relying on active contours, graph cuts and convolutional neural networks. We applied these methods on mouse brain manganese enhanced MR images acquired at 100 micrometre isotropic resolution, in vivo. All three methods achieved Dice coefficients larger than 94%, but convolutional neural networks achieved a small but significant improvement (0.97±0.01) over our active contours implementation (0.94±0.05) and the difference approached significance relative to graph cuts (0.96±0.01).
INTRODUCTION
Skull stripping often provides the initial step in neuroimaging pipelines. The accuracy of brain extraction impacts downstream processing steps. Automatic brain extraction procedures are widely used in human neuroimaging1,2 while extracting brain from preclinical data remains understudied3. We compared the performance of three methods relying on morphological snakes5, graph cuts4, and convolutional neuronal networks for mouse brain in vivo manganese enhanced MRI (MEMRI).METHODS
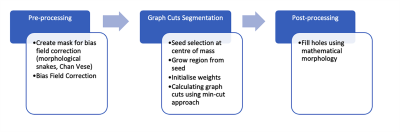
High resolution anatomical images are required for detecting volume changes in mouse models of aging, neurological conditions, or to compare function. We have imaged 33 mice, aged 12-18 months, 8 males, 25 females, of 4 different genotypes: expressing human APOE2 and APOE3 genes, familial Azheimer's disease mutations i.e. APPSwDI/mNos2−/− and the control mNos2−/−. In vivo imaging used a 7 T, 20-cm bore Bruker BioSpec, with Avance III console, a 72 mm volume excite coil, and a 4 channel surface receive coil. We used a RARE sequence with TE/TR 25ms/300 ms, 2 NEX, echo separation 6.2 ms, RARE factor 8, BW 100kHz. The image matrix was 200x180x100, and field of view 20x18x10mm. Images were reconstructed at 100µm x100µm x100µm. Manual labels were generated using ITK-SNAP (itksnap.org). We used N4BiasFieldCorrection6 with an initial mask generated using morphological snakes, i.e. the Chan-Vese model, implemented in the Scikit-image package7 (https://scikit-image.org/). This was seeded in the centre of mass of the image, and allowed to grow.To produce the final brain segmentation relying on the Chan Vese method we generated 10 instances of morphological active contours without edges, varying the parameters between (4, 0.5), and (0.5, 4), and thresholded the summed results at 90%. Mathematical morphology was used to fill in holes.
For a graph cuts segmentation we adopted the isometric partitioning approach4, which cuts narrowly connected regions from the foreground. We chose an initial seed in the centre of mass of the image, assumed to be in the mouse brain, and grew it using a conservative Chan Vese model, chosen so that the initial mask contains only brain voxels. Initial weights between the source (foreground, brain) and image voxels were set to infinity, and between sink (background, non-brain) and image voxels were set to 0. Weights between voxels were initialised in the 6-connected neighbourhood as $$w(i,j)=max(D(vi),D(vj))$$ where D(vi) and D(vj), are distances from vertices to the ROI. Further, we modify weights between voxels so that the cut should be through voxels with low intensity.
Our graph cuts method used the max-flow algorithm8 (PyMaxFlow package). We “filled in” remaining holes. The flowchart for this process is shown in Figure 1.
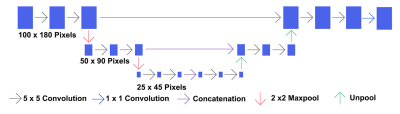
Lastly, we implemented a convolutional neural network (CNN) with a U-net architecture (Figure 2). This relied on TensorFlow 2.3 and consisted of 3 levels of 2 convolutional layers and a down-sampling layer, followed by 3 levels of 2 convolutional layers and an up-sampling layer. Each convolutional layer had a kernel of 5x5, unpadded, and we used rectified linear (ReLU) activation followed by a max-pooling layer to down-sample by 2x2. The final layer consists of a sigmoid activation that predicts whether a pixel belongs to the brain mask class or not. We optimized the model (1,487,169 parameters) using an Adam optimizer with exponential decay learning rate and binary cross entropy as a loss function. We used a leave one out strategy.
The skull stripping algorithms were validated against manually generated masks using Dice and Jaccard indices, sensitivity and specificity. We compared the three methods (active contours, graph cuts, and CNN using one way ANOVA, followed by Tukey post-hoc corrections.
RESULTS
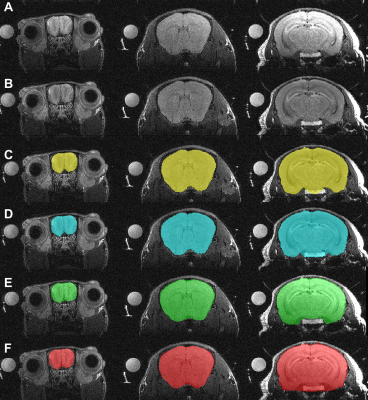
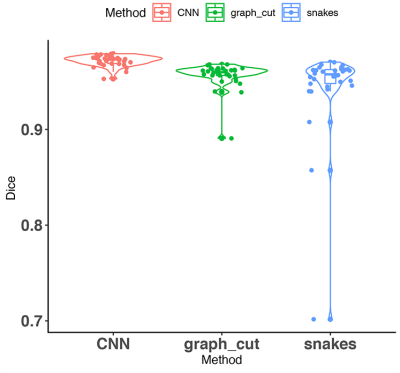
A qualitative comparison of unsupervised segmentation using the Chan Vese, graph cuts, and CNN brain segmentation, relative to the manual masks and the MR images is shown in Figure 3.A quantitative evaluation of the brain segmentation performance is presented in Figure 4. All methods produced average Dice coefficients > 94%. A one-way ANOVA revealed that differences, though small were significant (F(2,96)=7.135, p=0.001). The post-hoc tests revealed that the U-Net /CNN performed significantly better (0.97 ± 0.01) than the Chan-Vese/snakes model (adjusted p=0.001); and the difference between the U-Net/CNN and the graph cuts approached significance (adjusted p=0.088). The graph cuts approach resulted in Dice scores of 0.96 ± 0.01, while the snakes/Chan-Vese method results were 0.94 ± 0.04, but the difference was not significant.
Discussion
The unsupervised skull stripping methods did not outperform the deep learning approaches, but are fast (~200s using 12 core, AMDRyzen CPU), and can be readily used for preclinical data. The graph cuts method was more stable than the Chan-Vese approach, where some brains were segmented with ~0.70% accuracy. The CNN/U-Net required GPUs for efficient implementation and optimization, and was more time consuming (10 minutes using 4 RTX 8000 NVIDIA GPUs), but more accurate and stable, and thus shows promise for future multi-regional segmentation approaches.Conclusion
We have implemented three mouse brain segmentation methods for in vivo MEMRI. Unsupervised approaches can be readily incorporated into analysis pipelines, without needing sophisticated computers. The CNN approach was more accurate and more stable, but required training, which benefits from using GPUs. Trained models can be applied to future studies in animal models of neurological conditions, such as Alzheimer’s disease.Acknowledgements
This work was supported by the National Institutes of Health through R01 AG066184, RF1AG057895, R56 AG05789 and U24 CA220245. The study was also supported by the Research Centre for Informatics, grant number CZ.02.1.01/0.0/16~019/0000765 and by the grant Biomedical data acquisition, processing and visualization, number SGS19/171/OHK3/3T/13.References
1. Smith, S. M. Fast robust automated brain extraction. Hum. Brain Mapp. 17, 143–155 (2002).
2. Sadananthan, S. A., Zheng, W., Chee, M. W. L. & Zagorodnov, V. Skull stripping using graph cuts. NeuroImage 49, 225–239 (2010).
3. Hoyer, C., Gass, N., Weber-Fahr, W. & Sartorius, A. Advantages and Challenges of Small Animal Magnetic Resonance Imaging as a Translational Tool. Neuropsychobiology 69, 187–201 (2014).
4. Grady, L. & Schwartz, E. L. Isoperimetric graph partitioning for image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence 28, 469–475 (2006).
5. Chan, T. & Vese, L. An Active Contour Model without Edges. in Scale-Space Theories in Computer Vision (eds. Nielsen, M., Johansen, P., Olsen, O. F. & Weickert, J.) 141–151 (Springer, 1999). doi:10.1007/3-540-48236-9_13.
6. Tustison, N. J. et al. N4ITK: Improved N3 Bias Correction. IEEE Transactions on Medical Imaging 29, 1310–1320 (2010).
7. Marquez-Neila, P., Baumela, L. & Alvarez, L. A Morphological Approach to Curvature-Based Evolution of Curves and Surfaces. IEEE Trans. Pattern Anal. Mach. Intell. 36, 2–17 (2014).
8. Boykov, Y. & Kolmogorov, V. An experimental comparison of min-cut/max- flow algorithms for energy minimisation in vision. IEEE Trans. Pattern Anal. Machine Intell. 26, 1124–1137 (2004).
9. Chou, N., Wu, J., Bingren, J. B., Qiu, A. & Chuang, K. Robust Automatic Rodent Brain Extraction Using 3-D Pulse-Coupled Neural Networks (PCNN). IEEE Transactions on Image Processing 20, 2554–2564 (2011).
Figures