3507
Automated assessment of paramagnetic rim lesions in multiple sclerosis patients with 3T and 7T MP2RAGE1Signal Processing Laboratory (LTS5), École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 2Medical Image Analysis Laboratory, Center for Biomedical Imaging (CIBM), University of Lausanne, Lausanne, Switzerland, 3Radiology Department, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 4Translational Neuroradiology Section, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, United States, 5Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 6Department of Neurology, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 7Department of Neurology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 8Department of Neurology, Cliniques Universitaires Saint-Luc, Université Catholique de Louvain, Brussels, Belgium, 9Department of Neurology, Johns Hopkins University, Baltimore, MD, United States, 10Neurologic Clinic and Policlinic, Departments of Medicine, Clinical Research and Biomedical Engineering, University Hospital Basel and University of Basel, Basel, Switzerland, 11Translational Imaging in Neurology (ThINk) Basel, Department of Biomedical Engineering, University Hospital Basel and University of Basel, Basel, Switzerland, 12Research Center for Clinical Neuroimmunology and Neuroscience (RC2NB) Basel, University Hospital Basel and University of Basel, Basel, Switzerland
Synopsis
Paramagnetic rim lesions (PRL) in multiple sclerosis are chronic inflammatory lesions depicted in susceptibility-based MRI where high PRL lesion burden has been associated with a more aggressive disease course. As their visual detection is subjective and time-consuming, a convolutional neural network (RimNet) has recently been developed and applied to 3T susceptibility contrast MR images. In this work, we evaluate a unimodal RimNet architecture based on either the MP2RAGE uniform contrast or the concurrently obtained T1 map at both 3T and 7T. Results show that prediction improves considerably at 7T, suggesting that 7T MP2RAGE might be helpful for automatically identifying PRL.
INTRODUCTION
Paramagnetic rim lesions (PRLs) in multiple sclerosis (MS) patients are chronic active lesions characterized by an accumulation of iron microglia/microphages at their edges1. Recent studies have shown an association between a higher PRL burden and a more aggressive disease course2. However, PRL detection is challenging and usually performed visually on 3T or 7T susceptibility-based MR images3. Recently, we described an automated method for assessing PRLs at 3T (RimNet)4. RimNet is based on a convolutional neural network and takes as input 3D patches centered on candidate lesions. Its best model leverages a combination of 3D T2*-weighted segmented echo-planar imaging (segEPI) magnitude and phase images5, achieving a lesion-wise sensitivity and specificity of 71% and 95%, respectively, in a cohort of 124 MS patients. In this study, we developed a unimodal RimNet architecture based on either the magnetization prepared 2 rapid acquisition gradient echoes (MP2RAGE)6 uniform images (UNI) or T1-map series. Its performance was evaluated in a cross-validation procedure on 76 cases imaged at 3T and, in a separate testing set, on 20 cases imaged at 7T.METHODS
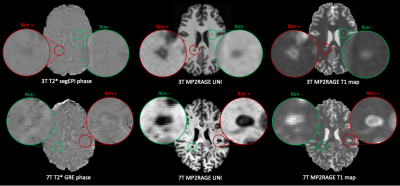
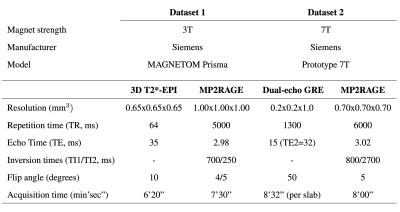
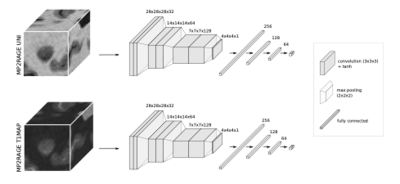
We retrospectively analyzed MRI data from 96 MS cases imaged at two university hospitals. Dataset-1 consisted of 76 cases (55 relapsing-remitting (RRMS) and 21 progressive MS (PMS), 49 female/27 male, mean age 41±14 years, age range [25-76] years, median EDSS 2.5, EDSS range [1.5-4.0]) imaged at 3T MRI (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany) using a 64-channel head coil. The protocol included MP2RAGE and 3D T2*-weighted segEPI, and the acquisition parameters are summarized in Table 1. Dataset-2 included 20 cases (13 RRMS/7 PMS, 15 female/5 male, mean age 44±11 years, age range [25-66] years, median EDSS 2.5, EDSS range [0-7.5]) scanned on a prototype 7T system (Siemens Healthcare, Erlangen, Germany) using a 32-channel head coil. The protocol, presented in Table 1, included MP2RAGE and T2*-weighted dual-echo gradient echo (GRE). In Dataset-1, MS lesions were automatically segmented7 and PRL manually annotated by two experts, who reached consensus in a joint session. An automated method optimized for 7T MP2RAGE8 was employed for segmenting MS lesions in Dataset-2, where a single expert visually identified PRL. Overall, Dataset-1 had a total of 2809 rim- lesions and 313 PRL, whereas Dataset-2 had 2308 rim- lesions and 212 PRL. MP2RAGE images were affinely registered to the T2* segEPI space using SimpleElastix9. Histogram matching10 was performed between 3T and 7T images for both UNI and T1 map, considering only the voxels inside the skull, in order to mitigate the intensity value differences. A unimodal RimNet architecture (see Figure 2 for more details), implemented as described in the original work, was trained first with the MP2RAGE uniform image (UNI) and then with the T1-map images from Dataset-1 in a four-fold nested cross-validation procedure. Inference was performed on both Dataset-1 and Dataset-2; for the former, an ensemble of the four trained classifiers was used. The areas under the ROC and PR curve (AUC), as well as the lesion-wise confusion matrices, were computed for performance evaluation.RESULTS
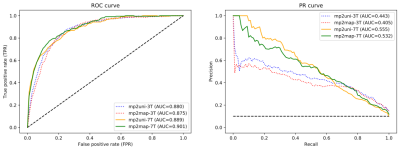
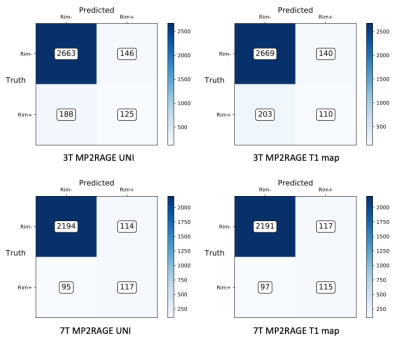
The ROC and PR curves, together with their relative AUC, are reported in Figure 3 for all four contrasts tested. The two models applied to 7T MP2RAGE UNI and T1-maps achieved the best results, with a PR AUC of 0.56 and 0.53, respectively. They clearly outperformed the 3T UNI and T1 map models, which reached AUC of 0.44 and 0.41. In Figure 4, we present the lesion-wise confusion matrices of the four models. By setting a false positive rate of 0.05, as in the original RimNet paper4, the top-performing model based on 7T UNI images achieved a sensitivity of 55%, specificity of 95%, positive predictive value of 50%, and negative predictive value of 96%.DISCUSSION
In this work, we explored the ability of a unimodal RimNet architecture based on either MP2RAGE uniform images or T1 maps to automatically assess PRL, at both 3T and 7T. Results show that, at both magnetic field strengths, MP2RAGE has some PRL detection capability, and that the UNI images slightly outperform the T1 maps. Moreover, even though RimNet was trained exclusively on 3T images, all evaluation metrics considerably improved for both images when tested at 7T. The best model, represented by the 7T MP2RAGE UNI, reached a PR AUC of 0.56, which is comparable to the AUC of 0.54 reported in the original RimNet paper for the unimodal T2*-w magnitude image at 3T, but still worse than the AUC for the phase image. This confirms, as we previously suggested4, that morphometric features such as size, shape, and signal intensity, all well depicted by MP2RAGE, are relevant in the classification of PRLs.CONCLUSION
Our contribution is two-fold. First, we show that the UNI and T1-map images of the MP2RAGE sequence acquired at 3T have some PRL prediction capability. Second, we observe that they considerably improve RimNet’s performance at 7T, suggesting that the MP2RAGE might be a suitable sequence for PRL assessment at 7T. Future work will explore integrating the MP2RAGE with the T2*-weighted magnitude and phase images, as well as the quantitative susceptibility mapping.Acknowledgements
This project is supported by the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie project TRABIT (agreement No 765148). The work is also supported by the Centre d'Imagerie BioMedicale (CIBM) of the University of Lausanne (UNIL), the Swiss Federal Institute of Technology Lausanne (EPFL), the University of Geneva (UniGe), the Centre Hospitalier Universitaire Vaudois (CHUV), the Hôpitaux Universitaires de Genève (HUG), and the Leenaards and Jeantet Foundations. CG is supported by the Swiss National Science Foundation grant PP00P3-176984.References
1. Absinta M, Sati P, Schindler M, et al. Persistent 7-tesla phase rim predicts poor outcome in new multiple sclerosis patient lesions. J Clin Invest. 2016; 126 (7): 2597-2609. doi: 10.1172 / JCI86198
2. Harrison DM, Li X, Liu H, et al. Lesion Heterogeneity on High-Field Susceptibility MRI Is Associated with Multiple Sclerosis Severity. Am J Neuroradiol. 2016; 37 (8): 1447-1453. doi: 10.3174 / ajnr.A4726
3. Kaunzner UW, Kang Y, Zhang S, et al. Quantitative susceptibility mapping identifies inflammation in a subset of chronic multiple sclerosis lesions. Brain. 2019; 142 (1): 133-145. doi: 10.1093 / brain / awy296
4. Barquero G, La Rosa F, Kebiri H, et al. RimNet: A deep 3D multimodal MRI architecture for paramagnetic rim lesion assessment in multiple sclerosis. NeuroImage Clin. 2020; 28: 102412. doi: 10.1016 / j.nicl.2020.102412
5. Sati P, Thomasson D, Li N, et al. Rapid, high-resolution, whole-brain, susceptibility-based MRI of multiple sclerosis. Mult Scler J. 2014; 20 (11): 1464-1470. doi: 10.1177 / 1352458514525868
6. Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele P-F, Gruetter R. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage. 2010; 49 (2): 1271-1281. doi: 10.1016 / j.neuroimage.2009.10.002
7. La Rosa F, Abdulkadir A, Fartaria MJ, et al. Multiple sclerosis cortical and WM lesion segmentation at 3T MRI: a deep learning method based on FLAIR and MP2RAGE. NeuroImage Clin. Published online June 2020: 102335. doi: 10.1016 / j.nicl.2020.102335
8. Fartaria MJ, Sati P, Todea A, et al. Automated Detection and Segmentation of Multiple Sclerosis Lesions Using Ultra-High-Field MP2RAGE: Invest Radiol. 2019; 54 (6): 356-364. doi: 10.1097 / RLI.0000000000000551
9. Marstal K, Berendsen F, Staring M, Klein S. SimpleElastix: A User-Friendly, Multi-lingual Library for Medical Image Registration. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW). ; 2016: 574-582. doi: 10.1109 / CVPRW.2016.78
10. van der Walt S, Schoenberger JL, Nunez-Iglesias J, et al. scikit-image: image processing in Python. PeerJ. 2014; 2: e453. doi: 10.7717 / peerj.453
Figures