3505
Rapid Estimation of Multiple Diffusion Maps from Undersampled Q-Space Data: A Comparison of Three Deep Learning Approaches1Radiology and Imaging Science, University of Utah, SALT LAKE CITY, UT, United States, 2Electrical and Computer Engineering, University of Utah, SALT LAKE CITY, UT, United States, 3Biomedical Engineering, University of Utah, SALT LAKE CITY, UT, United States
Synopsis
Advanced diffusion models enable characterization of tissue microstructure with higher specificity than conventional DTI. While beyond DTI diffusion imaging have been found valuable in many studies, their clinical availability have been hampered mainly due to their long scan times. Furthermore, each diffusion model can only extract a few relevant microstructural features. Therefore, using multiple models helps to better understand the brain microstructure, which requires multiple expensive model-fitting. In this study, we use different deep learning approaches to jointly estimate multiple advanced diffusion maps from highly undersampled q-space data, which can reduce both the scan and processing times significantly.
Introduction
Diffusion magnetic resonance imaging is a well-established technique in neuroimaging to probe microstructural and material properties of tissues non-invasively. While diffusion tensor imaging (DTI) is regularly used in clinical settings, more advanced diffusion representations and models such as diffusion kurtosis imaging (DKI)[1] , neurite orientation dispersion and density imaging (NODDI)[2], and multi-compartment spherical mean technique (SMT)[3] are not clinically available due to long acquisition times. Model-fitting can be computationally expensive, especially when multiple models are of interest.In this work, we study three deep learning (DL) approaches to estimate multiple diffusion parameters. Our main contributions are: (1) We estimate diffusion maps of four models simultaneously as opposed to prior works that estimate the models separately. (2) We train DL networks using HCP-A database and perform testing using locally acquired data of a stroke patient. This shows the trained models can be scanner and subject independent. (3) We study the impact of preprocessing on the performance of DL approaches and propose a more realistic approach of preprocessing raw DWIs after q-space undersampling. (4) Our results show that DL approaches can reduce both the scan and processing times significantly while maintaining small estimation errors of diffusion maps.
Materials and Methods
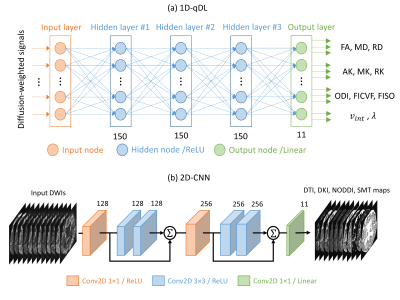
We compare three DL approaches: a per-voxel-based q-space deep learning (1D-qDL)[4], a per-slice-based convolutional neural network (2D-CNN)[5], and a 3D patch-based microstructure estimation with sparse coding using a separable dictionary (MESC-SD)[6] network. Network architectures are shown in Fig. 1 (a), Fig. 1 (b), and Fig. 2, respectively. Network inputs are q-space undersampled DWIs. Outputs are DTI ( fractional anisotropy(FA), mean diffusivity (MD), radial diffusivity (RD)), DKI ( axial kurtosis (AK), mean kurtosis (MK), radial kurtosis (RK)), NODDI ( orientation dispersion index (ODI), isotropic volume fraction ($$$v_{iso}$$$), intra-cellular volume fraction ($$$v_{ic}$$$)), and multi-compartment SMT maps ( intra-axonal volume fraction ($$$v_{int}$$$), intrinsic diffusivity ($$$\lambda$$$)).Networks were trained using diffusion datasets of 10 subjects 70-80 years old from the Human Connectome Project in Aging (HCP-A)[7]database. Target maps were computed using fully-sampled HCP-A diffusion acquisitions. Networks were tested using datasets of 15 subjects of the same age range from the HCP-A database. Also, a stroke patient with informed consent and IRB approval was imaged on a 3T Siemens Prisma scanner using the HCP-A diffusion protocol for testing.
For reference diffusion maps, prior to model fitting, DWIs were preprocessed using a pipeline performing ringing artifact removal[8], brain extraction[9,10], EPI susceptibility distortion correction [9,11], and eddy current and subject motion corrections [9,12]. For DTI, DKI, and NODDI we used diffusion imaging in Python (DIPY)[13] tools for model-fitting. For DTI/DKI, a weighted least-squares algorithm and for NODDI, accelerated microstructure imaging via convex optimization (AMICO) [14] were used. The multi-compartment SMT model-fitting was performed using a C++ compiled software provided by Kaden et al.[3].
Each network was trained for three q-space undersampling patterns with 60, 30, and 12 DWIs consisting of b=0, and b=1500 and 3000 s/mm2 shells. This reduces scan time from ~ 22 min to ~3.3 min, ~1.6 min, and 40 sec, respectively, and preprocessing time from ~5 hour to < 30 min. For each undersampling pattern, network training/testing was performed with two approaches: (1) preprocessed DWIs were undersampled in q-space. (2) raw DWIs were undersampled first and then preprocessed. We note that the first approach is not possible in practice, yet was used by all DL works to date [4-6]. The second approach simulates shortened acquisitions more realistically.
All networks were implemented in Python using Pytorch. For 1D-qDL, 4770934 training/validation samples (voxels) with a 95/5 split, 100 epochs, Adam optimizer ($$$\beta_1 =0.9$$$, and $$$\beta_2=0.999$$$), initial learning rate of 0.001, and L2 loss were used. Learning settings were the same for 2D-CNN and MESC-SD, except for 920 training/validation samples (slices) and L1 loss for 2D-CNN, and 10 epochs for MESC-SD. Using the same hardware resources, training/testing (per dataset) took ~1 hour/3-10 sec for 1D-qDL and 2D-CNN and ~10 hour/1min for MESC-SD. Using DL methods reduced the model-fitting time of 11 parameters from ~6 hours to < 1 min.
For quantitative evaluation of networks, the normalized root-mean-square-errors (nRMSE) of estimated diffusion maps were computed in 48 regions of interest (ROIs) in brain white matter for test datasets. The labels for ROIs were extracted by registering reference FA maps onto Johns Hopkins University FA atlas [15] using Advanced Normalization Tools (ANT)[16].
Results
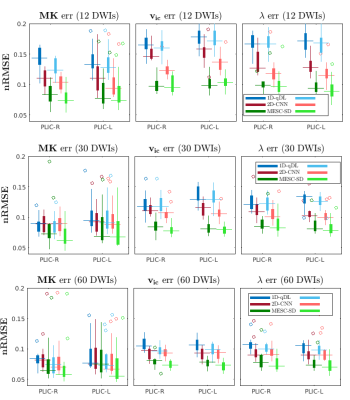
The nRMSE of three diffusion maps for 15 HCP-A database test subjects are shown in Fig. 3. The estimated diffusion maps and corresponding error images for the stroke patient test dataset are shown in Fig. 4 and Fig. 5, respectively.Conclusions
In this study, we compared three different DL approaches to estimate DTI, DKI, NODDI, and multi-compartment SMT maps from q-space undersampled DWIs. DL networks were trained using HCP-A database and tested using a stroke patient dataset acquired locally. We also studied impact of DWIs preprocessing on networks performances. The MESC-SD had the best performance among the three networks. The networks were able to estimate diffusion maps in the stroke regions with quality similar to other regions even though no stroke dataset was used for training. We found that DL approaches can reduce both the scan and processing times significantly while providing good estimates of multiple diffusion maps.Acknowledgements
The Prisma MRI scanner and MRI Recharge Center resources used were supported in part by the NIH Shared Instrumentation Grant S10OD018482. We thank Dr. Douglas Dean and Dr. Andrew Alexander for providing the diffusion preprocessing pipeline.References
[1] Jensen JH, et al. Diffusional kurtosis imaging: The quantification of non-Gaussian water diffusion by means of magnetic resonance imaging. Magnetic Resonance in Medicine, 2005.
[2] Zhang H, et al. NODDI: Practical invivo neurite orientation dispersion and density imaging of the human brain. NeuroImage, 2012.
[3] Kaden E, et al. Multi-compartment microscopic diffusion imaging. NeuroImage, 2016.
[4] Golkov V, et al. Q-space deep learning: Twelve-fold shorter and model-free diffusion MRI scans. IEEE Transactionson Medical Imaging, 2016.
[5] Gibbons EK, et al. Simultaneous NODDI and GFA parameter map generation from subsampled q-space imaging using deep learning. Magnetic Resonance in Medicine, 2019.
[6] Ye C, et al. An improved deep network for tissue microstructure estimation with uncertainty quantification. Medical Image Analysis, 2020.
[7] Harms MP, et al. Extending the Human Connectome Project across ages: Imaging protocols for the life span development and aging projects. NeuroImage, 2018.
[8] Kellner E, et al. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magnetic Resonance in Medicine, 2016.
[9] Jenkinson M, et al. FSL. NeuroImage, 2012.
[10] Smith SM. Fast robust automated brain extraction. Human Brain Mapping, 2002.
[11] Andersson JLR, et al. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage, 2003.
[12] Andersson JLR, et al. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage, 2016.
[13] Garyfallidis E, et al. Dipy, a library for the analysis of diffusion MRI data. Frontiers in Neuroinformatics, 2014.
[14] Daducci A, et al. Accelerated microstructure imaging via convex optimization (AMICO) from diffusion MRI data. NeuroImage, 2015.
[15] Mori S, et al. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage, 2008.
[16] Avants BB, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage, 2011.
Figures