3504
Volumetric assessment of patients with Glioblastoma by HUMBLe: Hierarchical 3D U-Net for MRI Brain Lesion segmentation
Yuval Buchsweiler1,2, Orna Aizenstein3,4, Felix Bokstein3,5,6, Idan Bressler1,2, Netanell Avisdris2,7, Deborah T. Blumenthal3,5, Dror Limon3,8, Dafna Ben Bashat2,3,6, and Moran Artzi2,3,6
1The Iby and Aladar Fleischman Faculty of Engineering, Tel Aviv University, Tel Aviv, Israel, 2Sagol Brain Institute, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 3Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel, 4Division of Radiology, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 5Neuro-Oncology Service, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 6Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel, 7School of computer science and engineering, Hebrew University of Jerusalem, Israel, Jerusalem, Israel, 8Division of Oncology, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel
1The Iby and Aladar Fleischman Faculty of Engineering, Tel Aviv University, Tel Aviv, Israel, 2Sagol Brain Institute, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 3Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel, 4Division of Radiology, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 5Neuro-Oncology Service, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 6Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel, 7School of computer science and engineering, Hebrew University of Jerusalem, Israel, Jerusalem, Israel, 8Division of Oncology, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel
Synopsis
Brain tumor segmentation is highly important for clinical management. We propose HUMBLe, a hierarchical 3D U-Net for MRI Brain Lesion segmentation architecture. HUMBLe breaks down the segmentation into its separate classes: enhancing tumor, edema, and necrotic classes, and uses a classifier to merge the different segmentation results into a final segmentation mask. Evaluation was performed on multi-parametric longitudinal local dataset, of patients with Glioblastoma. Segmentation results obtained by HUMBLe on our cohort improved DICE scores by 7%-16% for the different tumor components, compared to segmentation performed using 3D U-Net based architecture trained on BraTS2019 and our cohort.
Introduction
High-grade-glioma (HGG) is an aggressive type of malignant brain-tumor that grows rapidly, usually requires surgery and radio/chemo-therapy and has poor survival prognosis. Measurements of changes in tumor size are important for accurate diagnosis and follow-up. Currently, the most widely used criteria for therapy response assessment in those patients is the response assessment in neuro-oncology criteria, which relies on 2D manual measurements, and thus is less accurate and prone to human errors1. Volumetric measurement is preferable, however in clinical setup, manual volumetric segmentation of HGG is usually inapplicable as it is highly challenging, time consuming and user dependent.In the last few years, the use of deep-learning methods for brain-tumor segmentation has gained popularity, with a great number of studies conducted on the BraTS dataset composed of newly diagnosed patients2. Most of previously suggested methods classify the tumor into its clinically significant components using multi-class segmentation algorithms. However, applying those methods for patients' therapy response monitoring is highly challenging due to the treatment related variability in intensities, presence of surgical cavities and fuzzy tumor borders that appear in the clinical domain following tumor resection and chemo/radio/biological therapies.
In this study, we propose HUMBLe, a hierarchical 3D U-Net based deep-learning segmentation architecture that breaks down the segmentation into its separate classes and uses a classifier to merge the different segmentation results into a final segmentation mask.
Methods
Input Data- BraTS2019 Dataset: Composed of MRI data of 261 patients with Glioblastoma, annotated with manual segmentation of tumor area.
- Local clinical Dataset: Comprised of longitudinal MRI data of six patients with Glioblastoma (4 scans per patient, total of 24 scans), all following tumor resection and chemotherapy/radiotherapy. Manual segmentation of tumor area was done in a similar manner to BraTS dataset.
- Imaging Data: All cases were composed of multi-modal MRI, which includes T1 weighted image (WI), T1WI contrast-enhanced (T1WICE), T2WI and FLAIR modalities. Annotations were comprised of the Enhancing Tumor, Edema and Necrotic Tumor.
- Data Preprocessing: Included image co-registration, bias field correction, background removal, image cropping (brain region delineation) and image resizing.
Proposed method (HUMBLe)
A hierarchical deep-learning segmentation architecture (HUMBLe) was used (Figure 1). Each tumor component (Enhancing Tumor, Edema and Necrotic Tumor) was segmented individually, using a separate network, based on Isensee's et al. 3D U-Net3. Each network was trained with complete images with a batch size of 1 and a focal-DICE4 loss function. For each network, the ground truth segmentation mask was filtered to include only the desired class. The segmentation masks of the three separate networks were fused using a one-vs-rest Linear-SVM5 classifier to automatically select the value of each pixel in the final segmentation mask.
Data Splitting
BraTS2019 training dataset was split into 80% training and 20% validation datasets. The second stage of training was performed on our local clinical dataset using a leave one out method.
Experimental Scheme
- 3D U-Net BraTS Trained: Training the 3D U-Net based network over the BraTS2019 dataset, with all 4 modalities as input, while predicting all class labels. Applying inference on our local clinical dataset.
- 3D U-Net Transfer Learning: Training the 3D U-Net based network over the BraTS2019 dataset, with all 4 modalities as input, while predicting all class labels, followed by fine-tuning the model weights by training on our dataset.
- HUMBLe: Training the 3 pathways of the HUMBLe architecture, incorporating the Linear-SVM Classifier (Figure 1) over the BraTS2019 dataset, followed by fine-tuning the model weights by training on our dataset.
The network's results were compared using the DICE-Sorensen score for the three experimental schemes. For the HUMBLe architecture, we determined for each tumor class separately, which combination of modalities (T1WI, T1WICE, T2WI, FLAIR) results with the highest DICE scores (on our dataset).
Results
- Segmentation results for the 3 experimental schemes are given in Table 1. Best segmentation results were achieved by HUMBLe using all 4 modalities as input for the Enhancing and Necrotic Tumor classes, and only using the T2WI and FLAIR for the Edema class (Figure 1). The fused segmentation map of the model showed significant improvement in DICE scores with 7% for the Whole Tumor (Enhancing+Edema+Necrotic), 16% for the Tumor Core (Enhancing+Necrotic) and 15% for the Enhancing Tumor, in our cohort over the other two models (Table 1).
- Figure 2 demonstrates segmentation comparison for a patient with GBM. As can be seen, the best results were obtained by HUMBLe, which copes better with the challenge of tumor segmentation given the tumor intensity variabilities and fuzzy borders following therapies.
- Figure 3 shows an example of longitudinal segmentation result for a patient treated with Bevacizumab (between time points 2 and 3), which in many cases results in challenges to radiological therapy response assessment in those patients, due to the substantial change in tissue intensities6. The success of HUMBLe in tumor segmentation in those cases demonstrates the clinical potential of this tool.
Discussion
In this work we proposed the HUMBLe architecture for volumetric assessment of patients with Glioblastoma. HUMBLe achieved the best segmentation results in terms of DICE score for patients following tumor resection and chemo/radio/biological therapies and thus, it may be integrated in clinical practice for longitudinal therapy response assessment for patients with Glioblastoma.Acknowledgements
No acknowledgement found.References
- Wen, P.Y., et al., Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. Journal of clinical oncology, 2010. 28(11): p. 1963-1972.
- Bakas, S., et al., Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv preprint arXiv:1811.02629, 2018.
- Isensee, F., et al. Brain tumor segmentation and radiomics survival prediction: Contribution to the brats 2017 challenge. in International MICCAI Brainlesion Workshop. 2017. Springer.
- Abraham, N. and N.M. Khan. A novel focal tversky loss function with improved attention u-net for lesion segmentation. in 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). 2019. IEEE.
- Cortes, C. and V. Vapnik, Support-vector networks. Machine learning, 1995. 20(3): p. 273-297.
- Arevalo, O.D., et al., Assessment of glioblastoma response in the era of bevacizumab: longstanding and emergent challenges in the imaging evaluation of pseudoresponse. Frontiers in neurology, 2019. 10: p. 460.
Figures
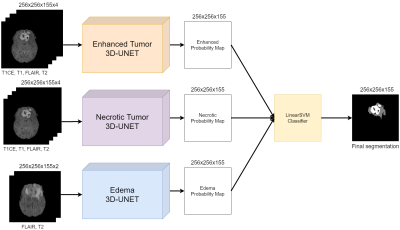
Figure 1: Hierarchihcal
3D U-Net architecture for MRI Brain Lesion segmentation (HUMBLe). The Enhancing and Necrotic Tumor pathways receive all 4 modalities as input while the Edema
receives only the Flair and T2WI.
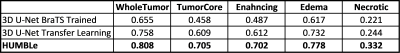
Table 1: Average
DICE scores for each tumor class and each architecture.
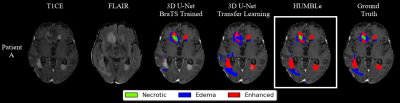
Figure 2: Architecture
segmentation comparison for a patient with GBM.
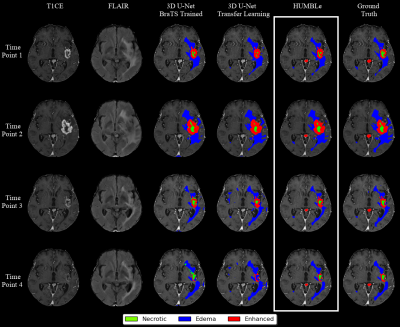
Figure 3: Architecture
segmentation comparison - Longitudinal volumetric assessment for a patient with
GBM