3499
Increasing Feature Sparsity in Alzheimer's Disease Classification with Relevance-Guided Deep Learning1Department of Neurology, Medical University of Graz, Graz, Austria
Synopsis
Using T1-weighted images we separated Alzheimer's patients (n=130) from healthy controls (n=375) by using a deep neural network and found that the preprocessing steps might introduce unwanted features to be used by the classifier. We systematically investigated the influence of registration and brain extraction on the learned features using a relevance map generator attached to the classification network. The results were compared to our relevance-guided training method. Relevance-guided training identifies sparser but substantially more relevant voxels, which improves the classification accuracy.
Introduction
Deep learning techniques are increasingly utilized in medical applications, including image reconstruction, segmentation, and classification.1,2 However, despite the good performance those models are not easily interpretable by humans.3 Especially medical applications require to verify that this is not the result of exploiting data artifacts.Our experiments on Alzheimer's disease (AD) classification showed that Deep Neural Networks (DNN) might learn from features introduced by the skull stripping algorithm. In this work we have investigated how preprocessing steps including registration and brain extraction determine which and how many features in the T1-weighted images are relevant for the separation of patients with AD from healthy controls.
Methodology
Dataset. We retrospectively selected 269 MRI datasets from 130 patients with probable AD (mean age=72.1±8.8 years) from our outpatient clinic and 484 MRIs from 375 age-matched healthy controls (mean age=66.7±10.5 years) from an ongoing community dwelling study. Patients and controls were scanned using a consistent MRI protocol at 3 Tesla (Siemens TimTrio) including a T1-weighted MPRAGE sequence with 1mm isotropic resolution. AD and HC data were split up randomly into five folds, while keeping all datasets from one person in the same fold. Overall folds were created by combining one fold from each cohort to ensure class distribution within.Preprocessing. Brain masks from each subject were obtained using BET from FSL. T1-weighted images were intensity normalized using a constant value and registered (A) affinely, using FSL flirt, and (B) nonlinearly, using FSL fnirt, to the MNI152 1mm template.4
Standard classification network. We utilized a classification network, combining a single convolutional layer followed by a down-convolutional layer as the main building block. The overall network stacks four of these main building blocks succeeded by two fully connected layers. Each layer is followed by a Rectified Linear Unit (ReLU) nonlinearity, except for the output layer where a Softmax activation is applied.
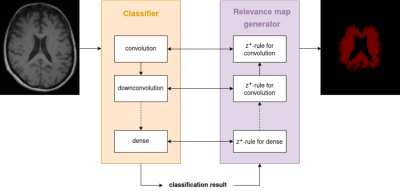
Relevance-guided classification network. To focus the network on relevant features, we proposed a relevance-guided network architecture, that extends the given classification network with a relevance map generator (cf. Figure 1 for details). To this end we implemented the deep Taylor decomposition (z+-rule) to generate the relevance maps of each input image depending on the classifier's current parameters.5
Training. We trained models for 3 differently preprocessed T1-weighted input images:
- in native subject T1-weighted image space
- linearly registered to MNI152 space
- nonlinearly registered to MNI152 space
Results
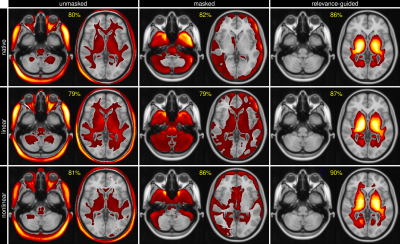
The classification accuracy between control subjects and AD was above 79% for all configurations, while the relevance-guided models showed substantially better results:- native space, unmasked / masked / relevance-guided: 80% / 82% / 86%
- linearly reg., unmasked / masked / relevance-guided: 79% / 79% / 87%
- nonlinearly reg., unmasked / masked / relevance-guided: 81% / 86% / 90%
Discussion and Conclusion
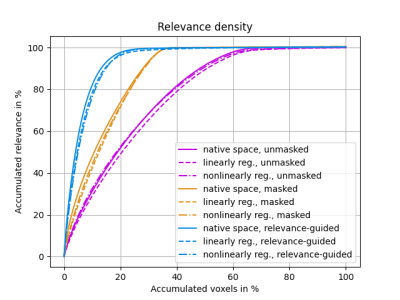
The results of this explorative study demonstrated that the preprocessing of MR images is crucial for the feature identification by DNNs. Previous work has shown that preprocessing may introduce features unrelated to the AD classification problem, e.g. results biased by brain extraction algorithms.7The proposed relevance-guided approach identified regions with highest relevance in brain tissue located adjacent to the ventricles. While AD progression is commonly paralleled by ventricular enlargement8,9, the individual contributions of volumetric features and T1-weighted contrast changes have to be further investigated.
In conclusion, the proposed relevance-guided approach is minimizing the impact of preprocessing by skull stripping and registration, rendering it a practically usable and robust method for DNN-based neuroimaging classification studies. Additionally, relevance-guiding forces the feature identification to focus on the parenchyma only and therefore provides physiological more plausible results.
Acknowledgements
This research was supported by NVIDIA GPU hardware grants.References
[1] Zhou T, Thung K-H, Zhu X, Shen D. Effective feature learning and fusion of multimodality data using stage-wise deep neural network for dementia diagnosis. Hum Brain Mapp. 2018.
[2] Liu S, Liu S, Cai W, Pujol S, Kikinis R, Feng D. Early diagnosis of Alzheimer’s disease with deep learning. 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI). IEEE. 2014; 1015–1018.
[3] Samek W, Binder A, Montavon G, Lapuschkin S, Muller K-R. Evaluating the Visualization of What a Deep Neural Network Has Learned. IEEE Trans Neural Netw Learning Syst. 2017;28: 2660–2673.
[4] Smith SM, Zhang Y, Jenkinson M, Chen J, Matthews PM, Federico A, et al. Accurate, Robust, and Automated Longitudinal and Cross Sectional Brain Change Analysis. Neuroimage. 2002;17: 479–489.
[5] Montavon G, Lapuschkin S, Binder A, Samek W, Müller K-R. Explaining nonlinear classification decisions with deep Taylor decomposition. Pattern Recognit. 2017;65: 211–222.
[6] Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. CoRR,vol. abs/1412.6980, 2014.
[7] Fennema-Notestine C, Ozyurt IB, et al. Quantitative Evaluation of Automated Skull-Stripping Methods Applied to Contemporary and Legacy Images: Effects of Diagnosis, Bias Correction, and Slice Location. Human Brain Mapp. 2006;27: 99-113.
[8] Apostolova LG, Green AE, Babakchanian S, et al.. Hippocampal atrophy and ventricular enlargement in normal aging, mild cognitive impairment and Alzheimer’s disease. Alzheimer Dis Assoc Disord. 2012;26(1): 17-27.
[9] Nestor SM, Rupsingh R, Borrie M, et al. Ventricular enlargement as a possible measure of Alzheimer’s disease progression validated using the Alzheimer’s disease neuroimaging initiative database. Brain. 2008;131(9): 2443–2454.
Figures