3495
Improving the generalizability of convolutional neural networks for T2-lesion segmentation of gliomas in the post-treatment setting1Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States, 2Center for Intelligent Imaging, UCSF, San Francisco, CA, United States
Synopsis
Routine monitoring of response to therapy in patients with glioma greatly benefits from using volumetrics quantified from lesion segmentation. Yet, the vast majority of deep learning models developed for this task have been trained using data from treatment-naïve, newly-diagnosed patients, whose T2-lesions have different appearance on imaging. We found that increasing the proportion of treated patients in training, incorporating a cross-entropy loss term that takes into account the spatial distance from surgical resection cavity and leading tumor edge, and transfer learning from newly-diagnosed to post-treatment imaging domains were effective strategies to improve the generalizability of segmentation of the T2-lesion post-treatment.
Introduction
Although current advances for automated glioma lesion segmentation and volumetric measurements using convolutional neural networks (CNN) have yielded high performance on newly-diagnosed patients1–3, there are challenges in generalizability for patients post-treatment4, where response criteria using these metrics are assessed routinely in the clinic. Surgical resections, adjuvant treatment, or significant disease progression, can greatly alter the characteristics of these lesions on T2-weighted imaging. To improve the generalizability of CNN segmentation to patients that have received prior treatment, we evaluated the effects of: 1) training on datasets with different combinations of newly-diagnosed and treated gliomas, 2) incorporating a loss term that takes into account the location of the surgical resection cavity and leading tumor edge, and 3) transfer learning to improve the segmentation accuracy of treated gliomas.Methods
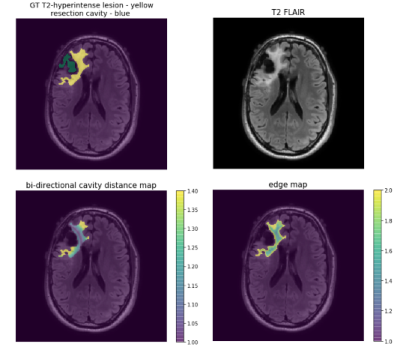
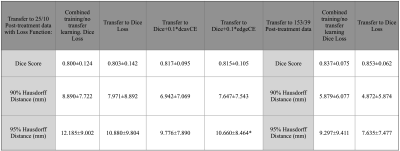
3D T2 FLAIR images (TE: 114ms-127ms, TR: 5850ms-6400ms, ETL:148-200, 1x1x1.5mm resolution) acquired on a 3T GE MR 750 scanner from 425 patients with glioma (208 newly-diagnosed 217 post-treatment) were used to train and evaluate modified versions of NVIDIA’s 2018 BraTS challenge winning variational autoencoder3 (VAE) to predict manually annotated T2-hyperintense lesions from a single input image. Training was first performed on 192 newly-diagnosed patients using NVIDIA’s Clara-Train software on a V100-32GB GPU. The proportion of training images from treated gliomas was then increased while maintaining 80%/20% train/validation splits. Models were tested on a separate set of 25 patients with post-treatment glioma. A second experiment was then performed with training/validation/test splits beginning with 153/39/25 volumes from treated patients and successively adding 50 newly-diagnosed patients into training/validation until all 208 were included. A subset of 25 post-treatment patients with annotated resection cavities was then used to compare the effects of using shape aware loss functions5 that introduce a cross entropy (CE) term to weight pixels according to their spatial distance to a boundary. This loss term was added to Dice loss and two VAE branch penalty terms, Kullback-Leibler (KL) divergence and L2 to form the final loss function: Ltot=λ1LDice+λ2LCE+λ3LKL+λ3LL2, where LCE=w(yi)*-∑i(yi)log(yi′)) with weightings of λ3,λ2=0.1, and λ1=1.0 and w(yi) is a generated weighting map based on shape. Two shape aware loss functions were evaluated and compared to Dice and VAE terms alone: 1) the positional relationship between the boundary of the cavity and T2 lesion (w(yi)=1+distcavity(yi)) or 2) uniform edge weighting from the T2 boundary (w(yi)=1+distedge(yi)) (Figure 1). Weighting maps were generated prior to training in order to minimize training times. These loss functions were first compared while training/validating/testing on 25/10/25 post-treatment patients before being used to finetune our pre-treatment model. To evaluate the effects of transfer learning as a strategy for generalization to images from treated glioma, we finetuned our pre-treatment model with both small (25/10) and large amounts (153/39) of post-treatment data. Models were evaluated based on mean Dice score and Hausdorff Distance (HD). 90th and 95th percentile cutoffs for HD were chosen to evaluate segmentation performance specifically at the lesion boundary while remaining robust to outlier predictions.Results & Discussion
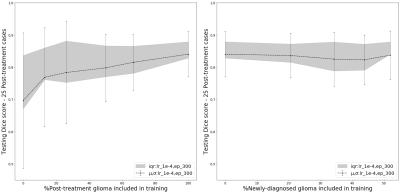
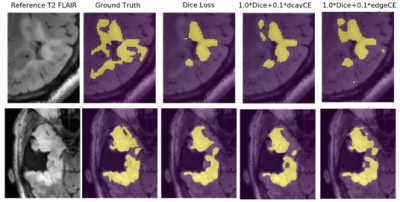
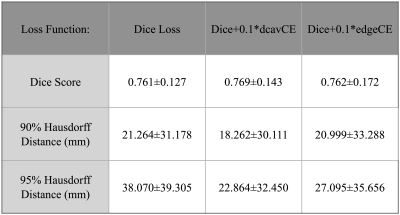
Including post-treatment scans as only 26% of the total training data substantially increased the mean testing Dice Scores by 12.5% after which a steady increase was observed to .840 when using 100% post-treatment data in training (Figure 2A). Models trained with additional images from newly-diagnosed gliomas experienced minimal change in testing Dice Scores, with the total combined model achieving a Dice Score of .837 (Figure 2B). Models trained and validated on the subset of 25/10 post-treatment patients with both edge and cavity distance weighted loss functions appeared to segment lesions containing small disconnected components in-plane more favorably than their counterparts trained without distance weightings (Figure 3). The model using the cavity loss term performed best in our evaluation metrics (Figure 4). When transfer learning was applied on a small subset of post-treatment patients, the models that were finetuned with distance weighted loss terms achieved higher performance on the 25 post-treatment test cases (Figure 5), compared to both finetuning with the standard loss function and training with all of the data without distance weighted loss terms. Incorporating a loss term that places additional emphasis on voxels adjacent to and far from the surgical cavity, or low-contrast voxels at edges of the lesion, improved segmentation performance in conventional training and transfer learning. Finetuning the newly-diagnosed pre-trained model with a nearly equal number of post-treatment images increased performance compared to the combined model (Figure 5), indicating improved generalization to the post-treatment imaging domain with this approach.Conclusion
We have demonstrated the effectiveness of three different training strategies for improving the generalizability of segmenting the T2-hyperintense lesion from T2 FLAIR images of treated gliomas. With limited post-treatment data, the best performance was achieved when pre-training with newly-diagnosed data followed by finetuning with post-treatment data using a loss function that incorporates a distance-weighted CE term based on proximity to the resection cavity. The overall best performance was obtained when employing transfer learning from pre- to post- treatment images with nearly equal amounts of patients. These results highlight the benefit of domain adaptation as a training strategy for increasing generalizability. Current efforts are underway to incorporate our models real time in the clinic for prospective validation and quantification of serial changes in tumor volumes.Acknowledgements
This work was supported by NIH NCI grant P01CA118816 and feasibility funding from HDFCC and cancer imaging resources (CIR).References
[1] Jiang, Z.; Ding, C.; Liu, M.; Tao, D. Two-Stage Cascaded U-Net: 1st Place Solution to BraTS Challenge 2019 Segmentation Task; 2020; pp 231–241. https://doi.org/10.1007/978-3-030-46640-4_22.[2] Kamnitsas, K.; Bai, W.; Ferrante, E.; McDonagh, S.; Sinclair, M.; Pawlowski, N.; Rajchl, M.; Lee, M.; Kainz, B.; Rueckert, D.; Glocker, B. Ensembles of Multiple Models and Architectures for Robust Brain Tumour Segmentation. ArXiv171101468 Cs 2017.
[3] Myronenko, A. 3D MRI Brain Tumor Segmentation Using Autoencoder Regularization. ArXiv181011654 Cs Q-Bio 2018.
[4] Zeng, K.; Bakas, S.; Sotiras, A.; Akbari, H.; Rozycki, M.; Rathore, S.; Pati, S.; Davatzikos, C. Segmentation of Gliomas in Pre-Operative and Post-Operative Multimodal Magnetic Resonance Imaging Volumes Based on a Hybrid Generative-Discriminative Framework. Brainlesion Glioma Mult. Scler. Stroke Trauma. Brain Inj. BrainLes Workshop 2016, 10154, 184–194. https://doi.org/10.1007/978-3-319-55524-9_18.
[5] Caliva, F.; Iriondo, C.; Martinez, A. M.; Majumdar, S.; Pedoia, V. Distance Map Loss Penalty Term for Semantic Segmentation. 5. 2019
Figures