3486
DL-BET - A deep learning based tool for automatic brain extraction from structural magnetic resonance images in mice.
Sabrina Gjerswold-Selleck1, Nanyan Zhu2,3,4, Haoran Sun1, Dipika Sikka1, Jie Shi1, Chen Liu3,4,5, Tal Nuriel4,6, Scott A. Small4,7,8, and Jia Guo3,9
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Biological Science, Columbia University, New York, NY, United States, 3Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States, 4Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, Columbia University, New York, NY, United States, 5Electrical Engineering, Columbia University, New York, NY, United States, 6Department of Pathology and Cell Biology, Columbia University, New York, NY, United States, 7Gertrude H. Sergievsky Center, Columbia University, New York, NY, United States, 8Radiology, Columbia University, New York, NY, United States, 9Psychiatry, Columbia University, New York, NY, United States
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Biological Science, Columbia University, New York, NY, United States, 3Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States, 4Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, Columbia University, New York, NY, United States, 5Electrical Engineering, Columbia University, New York, NY, United States, 6Department of Pathology and Cell Biology, Columbia University, New York, NY, United States, 7Gertrude H. Sergievsky Center, Columbia University, New York, NY, United States, 8Radiology, Columbia University, New York, NY, United States, 9Psychiatry, Columbia University, New York, NY, United States
Synopsis
Brain extraction plays an integral role in image processing pipelines in both human and small animal preclinical MRI studies. Due to lack of state-of-the-art tools for automated brain extraction in rodent research, this step is often performed semi-supervised with manual correction, making it prone to inconsistent results. Here, we perform a multi-model brain extraction study and present a semi-automated preprocessing workflow and deep neural network with a 3D Residual Attention U-Net architecture as the optimal network for automated skull-stripping in neuroimaging analysis pipelines, achieving a DICE score of 0.987 and accuracy of 99.7%.
Introduction
Brain extraction is a fundamental step in preclinical MRI studies, representing an integral part of processing pipelines in both human and small animal MRI. Also known as skull-stripping, this processing step refers to the identification of the brain within the MRI volume and subsequent removal of the skull and surrounding tissues from an image, leaving only the region of brain tissue for further neuroimaging analysis. While much literature on brain extraction aims to accomplish this task in humans, brain extraction still poses a great challenge in rodents, as most human brain extraction algorithms are not directly suitable for rodents due to notable physiological and size differences between human and small animal brains. Due to lack of state-of-the-art tools for automated brain extraction in rodent research, this step is often performed manually in a way that is not only time-consuming but also prone to inconsistent results due to high variability between annotators. Brain extraction has also been performed through edge-based and atlas-based methods. Deep learning, a subset of machine learning, has demonstrated great promise for fast and robust brain extraction, as previously shown in humans and mice1,2. Here, we offer a semi-automated preprocessing workflow and deep learning-based brain extraction tool implementing a Residual Attention U-Net architecture that has previously been used for tumor segmentation3, semantic segmentation of surgical instruments4, and cell segmentation5, which we applied and optimized for the purpose of brain extraction. In this work, we demonstrate that the Residual Attention U-Net with 3D inputs performs brain extraction in mice MRI scans with high visual and quantitative accuracy. Our work will be released as a publically available deep learning brain extraction toolkit (DL-BET).Methods
This study involved 200 healthy adult wild type C576J/BL male mice scanned at 3-48 months. MRI acquisitions were performed using the 2D T2-weighted TurboRARE sequence at 9.4T on a Bruker BioSpec 94/30 scanner (TR/TE = 3500/45, RARE factor = 8, 76 µm in-plane resolution, 450 µm slice thickness). Images were preprocessed with isotropic upsampling to 60 micron resolution and N4 bias field correction. Intensity normalization to the dynamic range of [0,1] was done and a train-validation-test ratio at 8:1:1 was applied to model training. Ground truth brain masks were generated using PCNN6 to and multi-rater manual correction. Our study employs 2D and 3D Residual Attention U-Net (RAU-Net) architectures. Model inputs are MRI scans of uniform dimensions, while outputs are predicted brain masks. The RAU-Net is an extension of the U-Net,7 with the addition of residual blocks8 and the attention gates9. As an example of a convolutional neural network (CNN), the U-Net extracts imaging features by utilizing local convolutions along the entire image or volume. The U-Net consists of several encoding layers across which the image dimension shrinks whereas the feature dimension increases so that compact high-level abstractions are generated along the process, and the same number of decoding layers to decipher these abstractions into image space information. The residual blocks are added to simplify entities to be approximated across each layer and therefore enable training of deeper networks, while the attention gates learn to differentially enhance or suppress specific regions in the feature maps so that the downstream outcomes better suit the desired task. DICE coefficient, Jaccard index, accuracy, sensitivity, and specificity were used to quantify performance of RAU-net models.Results
Quantitative evaluation demonstrates high similarity between predictions by all models and corresponding ground truth masks (Figure 3). To validate the success of our model and benefit of implementing an architecture with attention for accurate skull-stripping, 2D and 3D RAU-Nets were compared to the SkullNet, a 2D CNN-based architecture designed by De Feo et al, as a baseline and standard for successful network performance in this study [1]. Both our 2D and 3D models outperform the SkullNet, with our 3D model being the most successful of the three models across all quantitative metrics, with a DICE score of 0.987 and accuracy of 99.7%, demonstrating great similarity to the ground truth masks. Visually, the 3D RAU-Net prediction appears most similar to the ground truth mask and is notably much smoother than the SkullNet prediction from the same input scan (Figure 4).Discussion & Conclusion
Based on the results of our multi-model brain extraction study, we present a semi-automated preprocessing workflow and deep neural network with a 3D RAU-Net architecture as the optimal alternative to manual brain extraction and the most favorable network for automated skull-stripping in neuroimaging analysis pipelines. We first show that our 2D model outperforms the previously published 2D SkullNet architecture, improving the accuracy and producing a mask with a smoother surface. Next, we show that our 3D model supersedes both 2D architectures, likely due to the bias incurred by analyzing a naturally three-dimensional object slice by slice when using a 2D model for skull-stripping. In the future, we aim to improve our model to have the capability of predicting masks with small training sets by introducing on-the-fly augmentation, which would allow us to adapt our model to facilitate brain extraction of other species with limited dataset availability.Acknowledgements
This study was performed at the Zuckerman Mind Brain Behavior Institute at Columbia University and Columbia MR Research Center site.References
- R. De Feo, A. Shatillo, A. Sierra, J. Miguel Valverde, O. Gröhn, F. Giove, and J. Tohka, “Automated skull-stripping and segmentation with Multi-Task U-Net in large mouse brain MRI databases,” bioRxiv preprint, doi.org/10.1101/2020.02.25.964015, 2020.
- S. Roy, A. Knutsen, A. Korotcov, A. Bosomtwi, B. Dardzinski, J. A. Butman, and D. L. Pham, “A Deep Learning Framework for Brain Extraction in Humans and Animals with Traumatic Brain Injury,” 2018 IEEE 15th Internsation Symposium on Biomedical Imaging, 978-1-5386-3636-7, 2018.
- Q. Jin, Z Meng, C. Sun, L. Wei, and R. Su. “RA-UNet: A hybrid deep attention-aware network to extract liver and tumor in CT scans,” arXiv preprint, arXiv:1811.01328, 2018.
- Z.-L. Ni, G.-B. Bian, X.-H. Zou, Z.-G. Hou, X.-L. Xie, C. Wang, Zhou, Li Y.-J, R.-Q., and Z. Li, “Raunet: Residual attention u-net for semantic segmentation of cataract surgical instruments,” arXiv preprint, arXiv:1909.10360, 2019.
- N. Zhu, C. Liu., Z. Singer, T. Danino, A. Laine, and J. Guo. “ Segmentation with residual attention u-net and an edge-enhancement approach preserves cell shape features,” arXiv preprint, arXiv:2001.05548
- N. Chou, J. Wu, J. Bai Bingren, A. Qiu, and K.H. Chuang, “Robust automatic rodent brain extraction using 3-D pulse-coupled neural networks (PCNN), ” IEEE Trans Image Process. 20(9): pp. 2554-2564, 2019, doi:10.1109/TIP.2011.2126587.
- O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional Networks for Biomedical Image Segmentation,” arXiv preprint, arXiv:1505.04597, 2015.
- K. He, X. Zhang, S. Ren, and J. Sun. arXiv preprint, arXiv: “Deep Residual Learning for Image Recognition,” arXiv preprint, arXiv:1512.03385, 2015.
- O. Oktay, J. Schlemper, L. Le Folgoc, M.C.H. Lee, M. P. Heinrich, K. Misawa, K. Mori, S. G. McDonagh, N. Y. Hammerla, B. Kainz, B. Glocker, and D. Rueckert. “Attention u-net: Learning where to look for the pancreas,” arXiv preprint, arXiv:1804.03999, 2018.
Figures
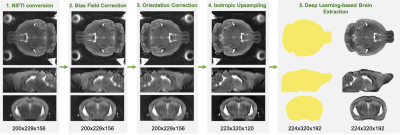
Preprocessing and Application Pipeline. Raw data is first converted to the NIFTI file type, followed by bias field correction, manual orientation correction, and isotropic upsampling. The preprocessing scan can then be fed into the deep learning-based brain extraction model to obtain an accurate brain mask.
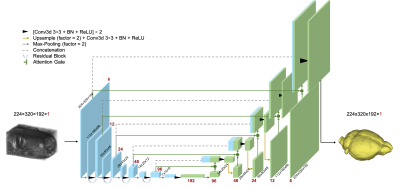
3D Residual Attention U-Net Architecture. The input of this network is a 3D MRI scan and the output is the corresponding 3D brain mask. 3D convolution and max-pooling are used in addition to residual blocks and attention gates. The function of each layer is defined in the legend.
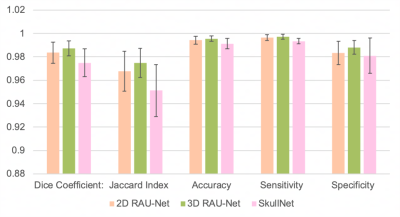
Quantitative performance comparison between three models based on five metrics: DICE coefficient, Jaccard index, accuracy, sensitivity, and specificity. Bars represent the mean of 20 test samples +/- standard deviation. All metrics are out of 1. Demonstrating the best performance across all metrics is the 3D Residual Attention U-Net, followed by the 2D Residual Attention U-Net.
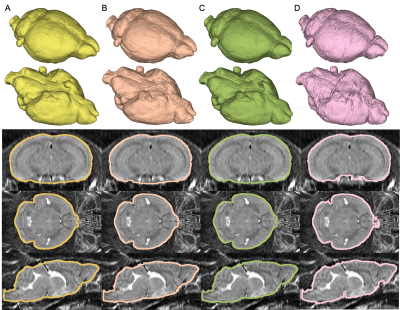
3D renderings and axial, coronal, and sagittal cross-section views of test predictions generated with the 2D RAU-Net (B), 3D RAU-Net (C), and SkullNet (D) compared to ground truth masks (A). A test subject was chosen that represented the best results of all three models, with masks having DICE coefficients of 0.991, 0.992, and 0.985 respectively.