3483
MRI based IDH and grade prediction using convolutional neural networks1Symbiosis centre for medical image analysis, Symbiosis International University, Pune, India, 2Dept of Radiology, National Institute of Mental Health and Neurosciences, Bengaluru, India
Synopsis
Recent developments in glioma subtyping suggest that IDH genotype as well as the histological grading are both crucial factors. However, earlier classification studies based on MRI features have focused either only on grade or IDH. In this work we employ an automated deep learning based technique to delineate the grade as well as the IDH status on a dataset of 178 subjects. Our classifier performs with a superior accuracy of 93.5% and the model explanability is achieved through class activation maps that illustrate the areas important in the classification.
Introduction
Based on 2016 World Health Organization (WHO) classification, gliomas are sub-categorized based not only on the histopathologic appearance but also on well–established molecular parameters such as the presence of IDH mutation and 1p19q codeletion1. Astrocytoma grade 2 and 3 have been classified in to IDH wild and mutant types while Glioblastomas which are grade 4 tumors are also classified in to IDH mutant and wild types. With improved understanding of glioma genetics, it is being recognized that Grade 2 and 3 IDH wild type gliomas have poor prognosis similar to the Grade 4 IDH wild type glioblastomas, while Grade 4 IDH mutant Glioblastoma are biologically distinct from the IDH wild type Glioblastoma and prognostically appear closer to the grade 2 and 3 IDH mutant tumors. These observations have been highlighted in the recent cIMPACT consortium2 recommendations. Additional biological markers are being identified to predict the biological behaviour of these tumors. However, currently both grading molecular characteristics like IDH mutations are being used for classification of Gliomas. Despite these developments earlier Glioma studies using quantitative MRI based analysis such as radiomics or convolutional neural nets (CNNs) have focused only on a single aspect – either grade or IDH mutation3. In this work we propose a CNN based model to classify the grade as well as IDH status for efficient delineation into WHO based categories.Methods
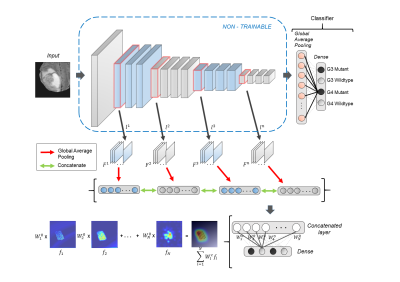
We combined a local dataset of 58 scans with 120 scans from The Cancer Genome Atlas (TCGA) dataset. This included 71 G3 scans (56 IDH-mutant, 15 IDH-wildtype) and 107 G4 scans (12 mutant, 95 wildtype). Our dataset included 24 adult patients (age = 48.8 ± 15.76 yrs, M: F = 11:13) with grade-4 gliomas and 34 adult patients (age 38.87 ± 10.98, M:F 21:13) with grade-3 gliomas (confirmed via histology). Multiple sequences were acquired as standard clinical MRI (on Philips 3T and Siemens 3T) however, we restricted our analysis to fluid attenuation inversion recovery imaging (FLAIR: TR/TE/T1= 11000/125/2800 ms within plane resolution of 0.5x0.5mm). Pre-processing of MRI images included: co-registration of all modalities to T1ce using six-degree rigid body transformation, resampling and transforming images to MNI space for a common origin, brain extraction using FSL-BET, manual correction and intensity normalization.A deep-learning based model was employed to distinguish between Grade3 and Grade4 glioblastomas along with their IDH status based on 2D FLAIR images. VGG-16, a sixteen-layer convolutional neural network (CNN) architecture4 was trained from scratch on boxed regions around tumors to distinguish between four classes namely G3-mutant, G3-wildtype, G4-mutant and G4-wildtype. The convolutional operation in initial layers of a CNN captures primitive features such as edges and curves from the image while the latter layers learn to encapsulate complex features such as shapes and patterns yielding prediction probabilities for every class. To increase the robustness of the model, augmentation methods involving rotating, flipping, shifting along height and width were used while training. Furthermore, to gain insights about the most discriminative regions in the tumor slices, we plot activation maps using HR-CAMs5 by sampling pre-maxpool convolutional layers. Weighted aggregation of the feature maps from the sampled layers results in precise localization of the most distinguishing regions in the image.
Results
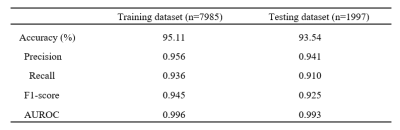
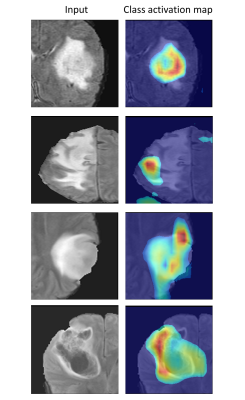
VGG-16 model performed with a normalized accuracy of 93.54% and balanced class accuracy of 91.06%. The classifier was able to differentiate between the 4 classes with average precision score of 0.941 and average recall score of 0.910 across all classes. Area under receiver operating characteristic curve (AUROC) was calculated with one-vs-rest approach and was found to be 0.993 for the test dataset as shown in fig 2. Fig 3 illustrates the heat-maps, that shows the most discriminative region for each image under consideration. The red area is highly weighted by CNN while blue being the least weighted.Conclusion
Our study focuses on the classification of high grade gliomas based on the grade as well as their IDH status. We demonstrate the feasibility to deploy an end-to-end deep learning model for precise delineation of the above mentioned tumor classes. Class activation maps provide significant insights about the areas on which the classification model focuses while giving its prediction probability for every class. This framework can potentially support pre-operative clinical prognosis and aid in treatment planning.Acknowledgements
No acknowledgement found.References
1. Louis, David N., et al. "The 2016 World Health Organization classification of tumors of the central nervous system: a summary." Acta neuropathologica 131.6 (2016): 803-820.
2. Louis, David N., et al. "cIMPACT‐NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT‐Utrecht meeting on future CNS tumor classification and grading." Brain Pathology (2020).
3. Chougule, Tanay, et al. "On Validating Multimodal MRI Based Stratification of IDH Genotype in High Grade Gliomas Using CNNs and Its Comparison to Radiomics." International Workshop on Radiomics and Radiogenomics in Neuro-oncology. Springer, Cham, 2019.
4. Simonyan, Karen, and Andrew Zisserman. "Very deep convolutional networks for large-scale image recognition." arXiv preprint arXiv:1409.1556 (2014).
5. Shinde, Sumeet, et al. "HR-CAM: Precise Localization of pathology using multi-level learning in CNNs." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2019.
Figures